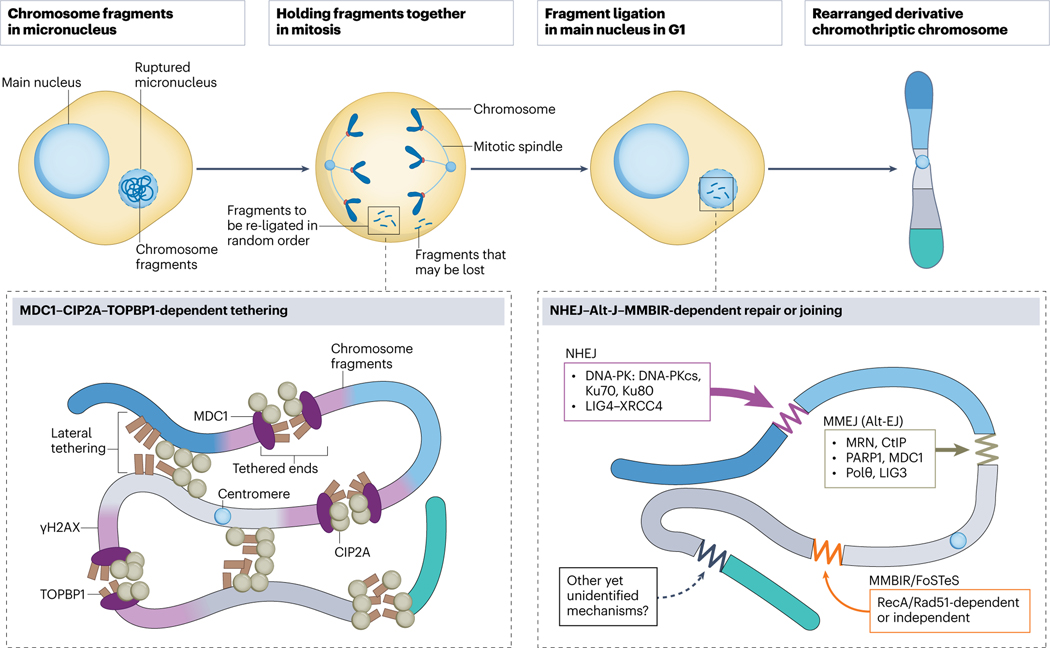
Fig. 4. Molecular mechanisms for chromosome reassembly.
Chromosome fragments from a ruptured micronucleus are held together in mitosis, which prevents their loss and is crucial for the subsequent reassembly of a derivative chromothriptic chromosome. This tethering is mediated by a three-protein complex which includes Mediator of DNA damage checkpoint 1 (MDC1) directly binding to γH2AX at double strand DNA breaks, Cellular Inhibitor of Protein phosphatase 2A (CIP2A) and DNA Topoisomerase II Binding Protein 1 (TOPBP1) or a two-protein complex of just CIP2A and TOPBP1. Reassembly of the random fragments into a chromothriptic chromosome occurs in a main nucleus in the next G1 phase. Non-homologous end-joining (NHEJ) is considered a major pathway involved in fragment re-ligation, followed by Microhomology-mediated end-joining, MMEJ (a type of alternative end-joining, Alt-EJ) which repairs clusters of DNA double strand breaks. Microhomology-mediated break-induced replication (MMBIR) and fork stalling and template switching (FoSTeS) are other replication-associated pathways which rely on microhomology.