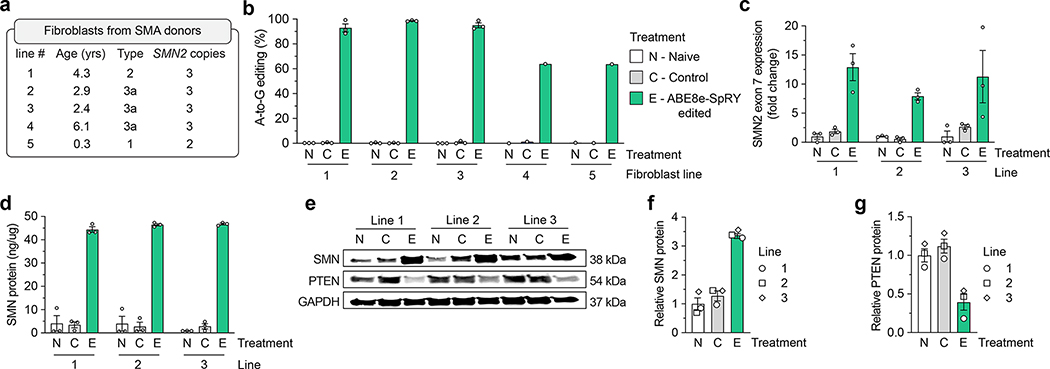
Figure 2. SMN2 C6T editing in SMA patient-derived fibroblasts.
a, Characteristics of five different SMA donors; all lines harbor a homozygous deletion of exon 7 in SMN1. b, A-to-G editing of the C6T adenine in SMN2 exon 7 across five SMA fibroblast cell lines transfected with ABE8e-SpRY and gRNA A8, assessed by targeted sequencing. Naïve (N) cells were untransfected; Control (C) cells were treated with ABE8e-SpRY and a non-targeting gRNA. c, SMN2 exon 7 mRNA expression across three edited (E) SMA fibroblast lines, measured by ddPCR. Transcript levels normalized by GAPDH mRNA, with data presented as normalized fold-change from naïve cells. d, SMN protein levels determined by an SMN-specific enzyme-linked immunosorbent assay (ELISA). e, Representative immunoblot for SMN, PTEN, and GAPDH protein levels across Naïve, Control, or ABE8e-SpRY treated SMA fibroblast lines. f,g, Quantification of SMN and PTEN (panels f and g, respectively) protein levels normalized to GAPDH and the Naïve treatment, determined by immunoblotting. For all assays, GFP-positive fibroblasts were sorted post-transfection and grown in for at least 3 passages; samples from three independent passages were collected for lines 1, 2 and 3 (passages 4–6; see Sup. Fig. 6a), and one passage was collected for lines 4 and 5. For panels b-d, f, and g, mean, s.e.m., and individual datapoints shown for n = 3 independent biological replicates from separate passages (unless otherwise indicated).