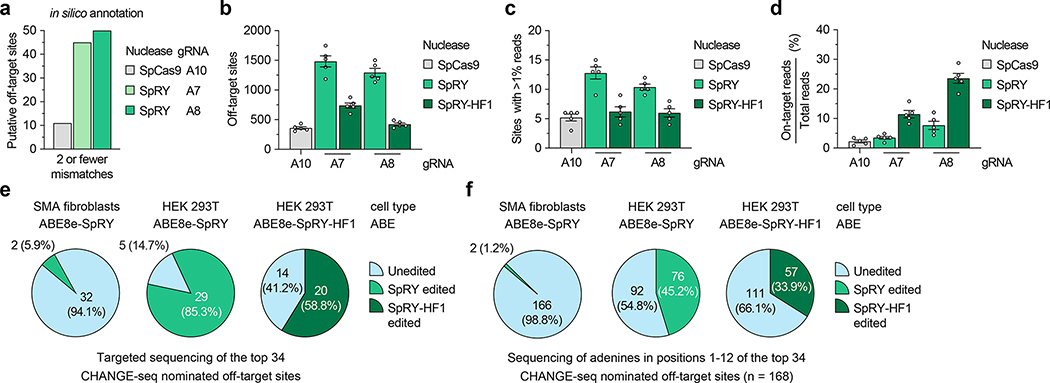
Figure 3. Analysis of SMN2 C6T base editing specificity.
a, Number of putative off-target sites in the human genome with up to 2 mismatches for each gRNA, annotated by CasOFFinder58. Predictions for SpCas9 utilized an NGG, NAG, or NGA PAM; with SpRY, a PAMless NNN search. b, Total number of CHANGE-seq detected off-target sites, irrespective of assay sequencing depth. c, Number of CHANGE-seq identified off-target sites that account for greater than 1% of total CHANGE-seq reads and are common across all 5 SMA fibroblast lines. d, Percentage of CHANGE-seq reads detected at the on-target site relative to the total number of reads in each experiment. For panels b, c, and d, mean, s.e.m, and individual datapoints shown for n = 5 independent biological replicate CHANGE-seq experiments (performed using genomic DNA from each of the 5 SMA fibroblast lines). e,f, Summary of targeted sequencing results from ABE-edited SMA fibroblasts or HEK 293T cells at the top 34 CHANGE-seq nominated off-target sites (common sites across all 5 SMA fibroblast lines and treatments with SpRY or SpRY-HF1), analyzing statistically significant editing of any adenine in the target site (panel e) or of all adenines in positions 1–12 of each of the 34 target sites (panel f).