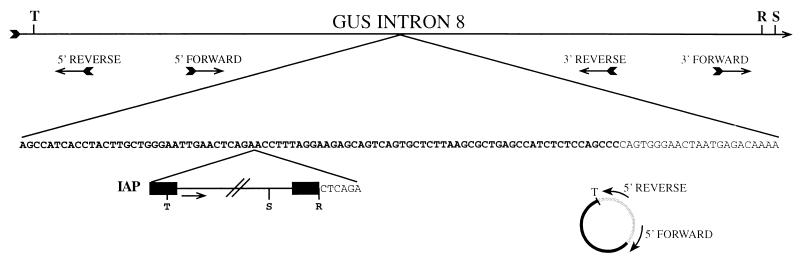
FIG. 4.
Schematic showing the insertion site of the IAP in intron 8 of the β-glucuronidase gene and the general method of inverse PCR cloning. The top line represents a fragment of intron 8, with the locations and orientations of the oligonucleotides indicated below; pertinent restriction enzyme sites are indicated above the line (R, RsaI; S, SspI; T, TaqI). Below, partial sequence of the intron shows the IAP integration site. Nucleotides in bold are part of the B2 repeat element. The remaining nucleotides represent nonrepetitive intron 8 sequence. The integrated IAP is represented at the bottom, flanked by its LTR sequences (black boxes). The direction of IAP transcription is indicated by the arrow. Restriction sites used in cloning are shown below the sequence. The six bases flanking the IAP represent the genomic sequence duplicated when the IAP integrated. An example of the mechanism by which insert sequence was obtained by inverse PCR is represented below the IAP. The circularization of the TaqI fragment at the 5′ end of the IAP element is shown. Intron 8-derived sequences are represented by the thin line, while those derived from the IAP LTR are indicated by the heavy line. Digestion of genomic DNA with TaqI cuts at the site adjacent to the oligonucleotide 5′ REVERSE (see cartoon above) and again at the site indicated in the 5′ LTR of the IAP. Subsequent circularization of that fragment via ligation produces the molecule shown, which includes sequences complementary to both 5′ REVERSE and 5′ FORWARD. Note that circularization places these oligonucleotides closer together and in an orientation suitable for PCR.