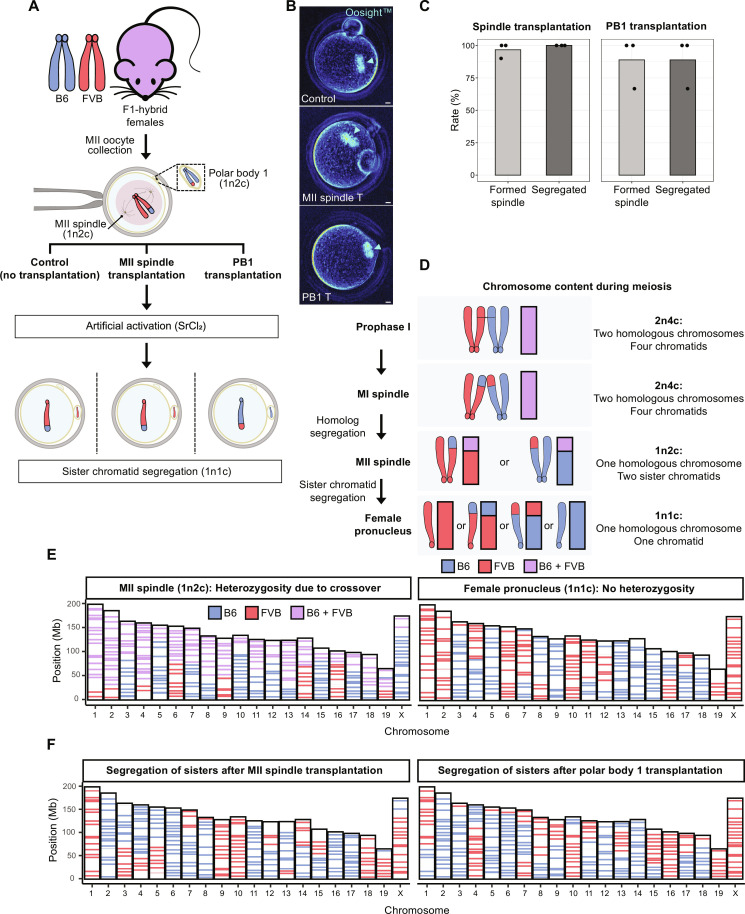
Fig. 1. MII cytoplasts support partitioning of transplanted 1n2c genomes by segregation of sister chromatids.
(A) Schematic of experiments investigating segregation of transplanted 1n2c genomes. (B) Spindle formation visualized using noninvasive Oosight live imaging. Control MII oocyte (top) with typical MII spindle pointed by arrowhead. Transplanted MII spindles (middle) or PB1 (bottom) formed typical spindles 2 hours after transplantation. Scale bar, 10 μm. (C) Spindle formation and genome segregation rates after MII spindle and PB1 transfer. Bar plots show the mean of three independent experiments (total n = 23 for MII spindle transplantation; total n = 7 for PB1 transplantation). (D) Illustration of chromosome ploidy (n) and chromatid number (c) at various stages of meiosis. (E) Analysis of chromosome origin and segregation in control, intact MII oocytes, and zygotes from B6/FVB F1 hybrid females via single-cell AmpliSeq sequencing. Left: Sequencing of DNA from MII spindles consisting of single set of chromosomes with two sister chromatids (1n2c). Right: Typical AmpliSeq profile for a haploid female pronucleus (1n1c) in zygotes after artificial activation of hybrid MII oocytes. (F) Analysis of chromosome segregation in zygotes after MII or PB1 transplantation. Left: Female pronucleus after transplantation of MII spindles. Right: Female pronucleus after transplantation of PB1. Note alternating homozygous B6 and FVB regions in individual chromosomes specific to 1n1c genomes.