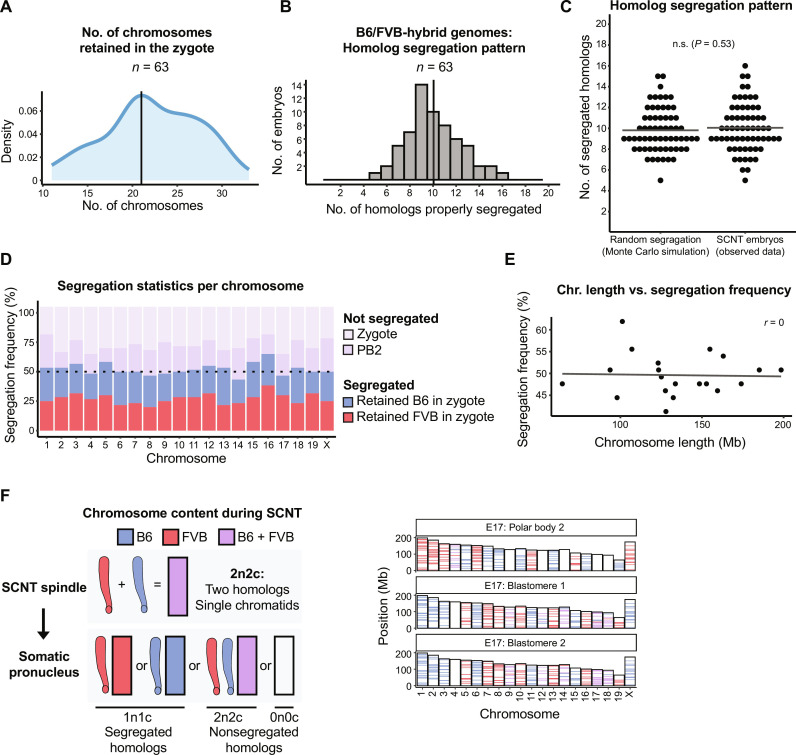
Fig. 3. Random chromosome segregation after premature division of diploid, nonreplicated 2n2c hybrid genomes.
(A) Number of chromosomes retained within a zygote upon partitioning of diploid, nonreplicated (2n2c) genomes in SCNT oocytes generated from hybrid B6/FVB somatic cell nuclei. Vertical line represents the mode number (21) derived from analysis of 63 SCNT embryos collected from eight independent experiments. (B) Number of homologous chromosomes properly segregated in 63 individual SCNT embryos. Vertical line indicates the mean number (10) of properly segregated homologs. (C) Comparison of homolog segregation patterns observed in SCNT embryos with random distribution expected from the Monte Carlo simulation. Horizontal lines represent the mean; n.s., not significant (P = 0.53, Wilcoxon rank sum test). (D) Segregation and nonsegregation statistics in SCNT embryos per individual mouse homolog. Horizontal dash line illustrates a hypothetical 50% chance for a pair of homologs to segregate (random segregation). (E) No linear relationship seen between chromosome length and homolog segregation frequency; r, Pearson correlation coefficient. (F) Illustration of the chromosome content (left) and segregation pattern in the SCNT embryo no. 17 (right) assessed by AmpliSeq analysis of DNA isolated from the PB2 and both blastomeres (trio). Only SCNT embryos producing sequence reads for all chromosomes in all trio samples (PB2 and two sister blastomeres) were included in analyses. Typical B6 or FVB sequence across all chromosomal regions in SCNT samples indicates absence of crossover recombination.