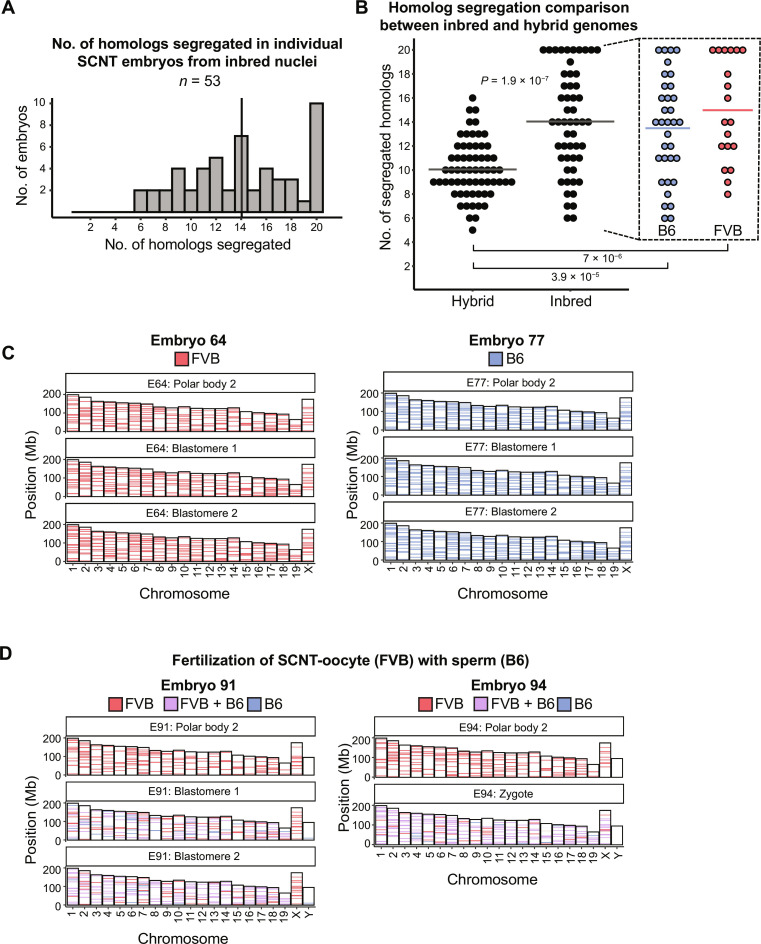
Fig. 4. Partitioning of diploid, nonreplicated 2n2c chromosomes from inbred genetic background results in haploid chromosome composition in daughter cells.
(A) Number of homologous chromosomes properly segregated in individual SCNT embryos (n = 53) composed of inbred FVB or B6 genomes. Vertical line indicates the mean (14). (B) Comparison of homologous chromosome segregation patterns between SCNT embryos composed of inbred (n = 53) versus hybrid (n = 63) somatic cell nuclei. Horizontal lines represent the mean; P = 1.85 × 10−7. Statistical significance was assessed by nonparametric, Wilcoxon rank-sum test. (C) Representative AmpliSeq chromosome sequencing data generated for the SCNT embryo no. 64 (left) and no. 77 (right) generated from FVB and B6 nuclei, respectively. (D) AmpliSeq chromosome sequencing profiles for two fertilized SCNT embryos showing proper segregation of all 20 homologs.