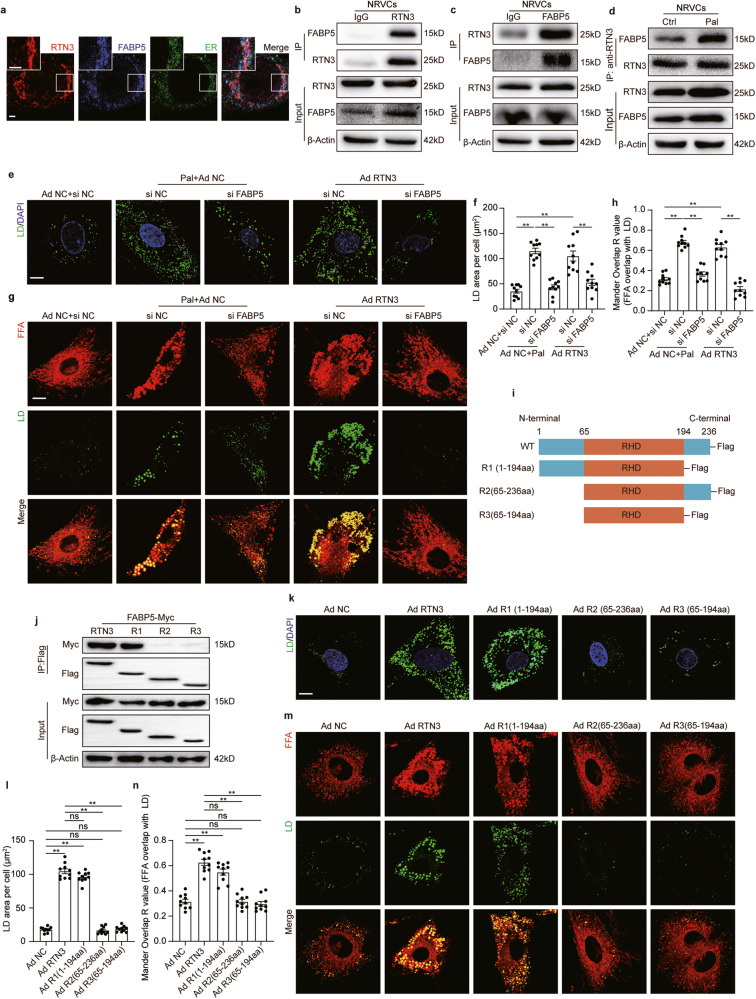
Fig. 5. RTN3 formed a protein complex with FABP5 to promote LD biogenesis.
a Representative immunofluorescence images indicating RTN3, FABP5, and ER, scale bar = 2.5 μm; Representative western blotting images of IP assay using anti-RTN3 (b) or anti-FABP5 (c) to determine the interaction of RTN3 and FABP5 in NRVCs; d Representative images of IP assay using anti-RTN3, NRVCs were used to perform the IP assay; Representative fluorescence images indicating LDs labeled with Bodipy 493/503 and nucleus labeled with DAPI (e) and quantitative analysis of LDs area per cell (f) (n = 10 images each group), scale bar = 10 μm; Representative fluorescence images indicating FFA labeled with Bodipy 558/568 C12 and LDs labeled with Bodipy 493/503 (g) and quantitative analysis of localization of FFA with LDs quantified with Mandar overlap R value analysis (h) (n = 10 images each group), scale bar = 10 μm; i Schematic diagrams of wild type and truncated forms of RTN3; j Representative western blotting images of IP assay using anti-Flag; Representative fluorescence images indicating LDs labeled with Bodipy 493/503 and nucleus labeled with DAPI in NRVCs infected with truncated RTN3 (k) and quantitative analysis of LDs area per cell (l) (n = 10 images each group), scale bar = 10 μm; Representative fluorescence images indicating FFA labeled with Bodipy 558/568 C12 and LDs labeled with Bodipy 493/503 in NRVCs infected with truncated RTN3 (m) and quantitative analysis of localization of FFA with LDs quantified with Mandar overlap R value analysis (n) (n = 10 images each group), scale bar=10μm. Data are expressed as Mean ± SEM. Differences are significant for **P < 0.01.