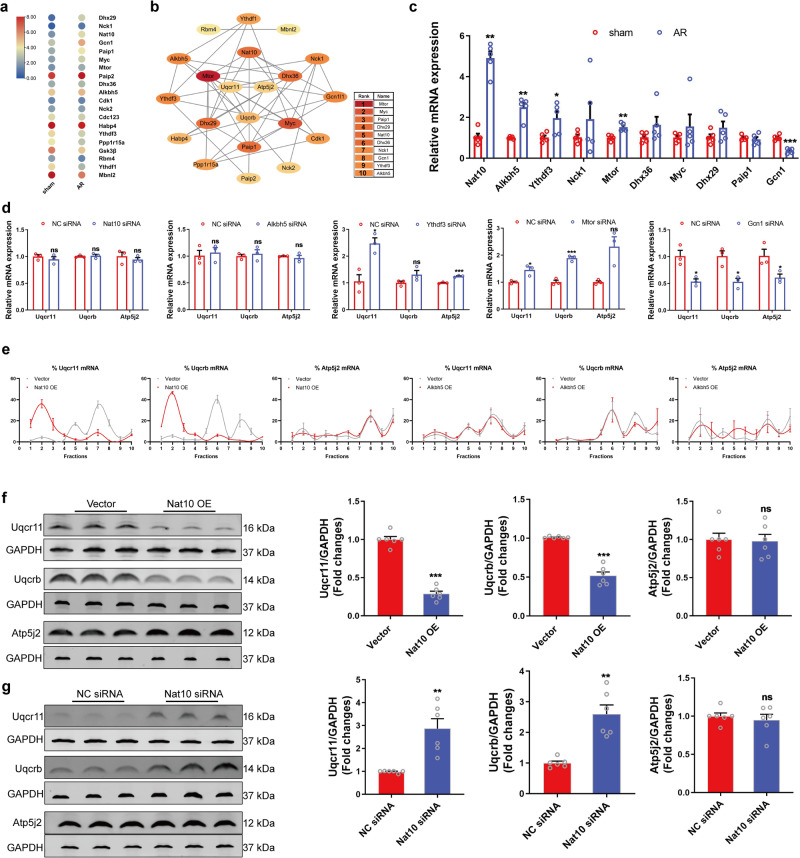
Fig. 3. The regulatory role of Nat10 in the translation of OXPHOS-related genes.
a Heatmap of genes involved in translational regulation in the RNA-seq data of AR mouse hearts. b A interaction network was constructed using STRING and visualized with Cytoscape (v. 3.7.1). c The expression of translation-related genes measured by qRT-PCR (n = 5 independent mice). *p < 0.05, **p < 0.01, ***p < 0.001 vs. sham group. d The effects of Nat10, Alkbh5, Paip2, Mtor, and Gcn1 on the mRNA expression of Uqcrb, Uqcr11 and Atp5j2 measured by qRT-PCR (n = 3 independent experiments). *p < 0.05, ***p < 0.001 vs. NC siRNA. e Polysome profiling analysis of Uqcrb, Uqcr11 and Atp5j2 in Nat10-overexpressed cardiomyocytes (n = 4 independent experiments). f and g Western blot analysis of Nat10, Uqcrb, Uqcr11 and Atp5j2 (n = 6 independent experiments). **p < 0.01, ***p < 0.001 vs. Vector or NC siRNA. Data were analyzed using Two-tailed Student’s t test with or without Welch’s correction for parameter analysis, Mann–Whitney two-tailed test for nonparametric analysis c, d, f, and g. Data are presented as mean ± SEM. Source data are provided as a Source Data file. Exact p values are provided in the Source Data file.