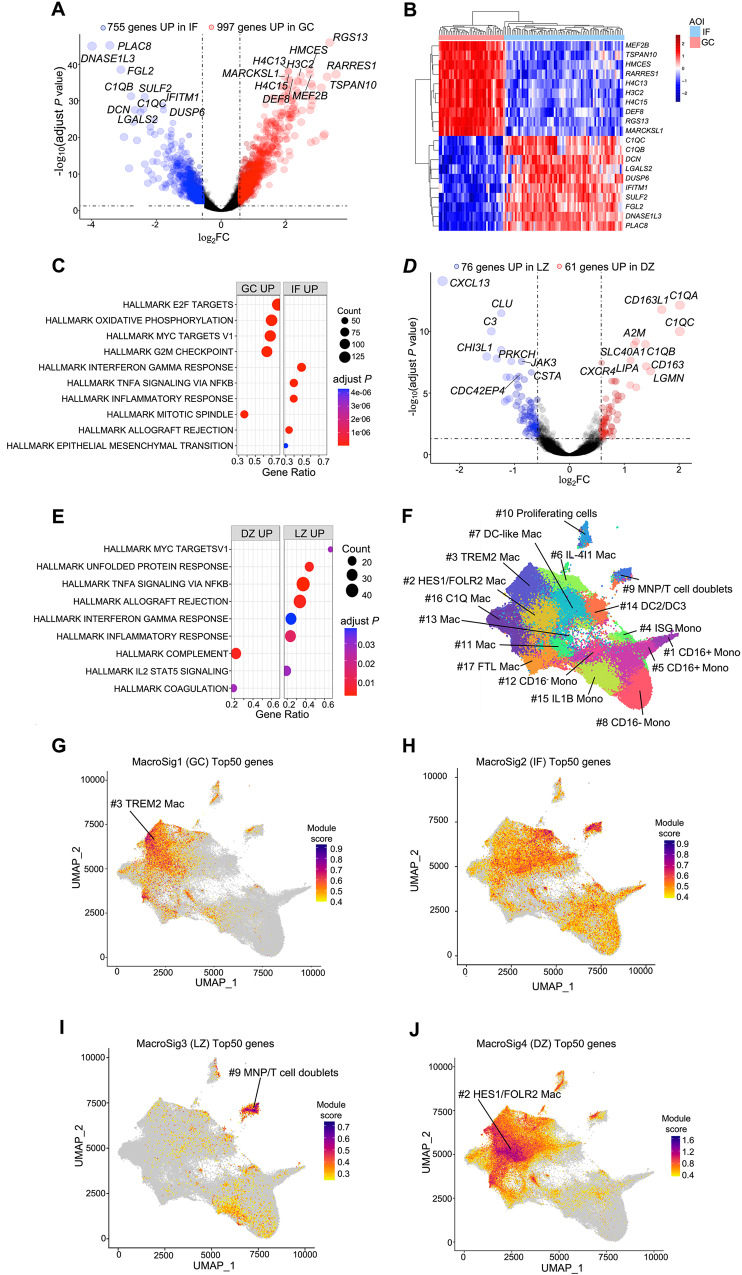
Fig. 2. Unique gene expression patterns differentiate macrophages in distinct spatial locations within reactive lymphoid tissues.
A Volcano plot showing the DEGs of macrophages between the GC and IF based on adjusted P < 0.05 and |log2FC| ≥ 0.58. P values were determined by two tailed moderated t test (BH corrected). B Top 20 macrophage DEGs (10 DEGs upregulated in GC and 10 DEGs upregulated in IF) are displayed based on adjusted P value in the heatmap. C Pathway enrichment analysis was performed on all DEGs between GC and IF. P value calculated by two tailed Fisher exact test (BH corrected). The top 10 pathways, based on BH adjusted P value, are shown. D The volcano plot showed the macrophage DEGs between LZ and DZ based on adjusted P < 0.05 and |log2FC| ≥ 0.58. P values were determined by two tailed moderated t test (BH corrected). E Pathway enrichment analysis was performed on all macrophage DEGs between LZ and DZ. P values were calculated by two tailed Fisher exact test (BH corrected). F MoMac-VERSE annotated 17 TAM subclusters using a compilation of 41 scRNA-seq datasets from 13 healthy and cancer tissues (Figure created via [https://macroverse.gustaveroussy.fr/2021_MoMac_VERSE/]). G–J Top50 genes of each MacroSig1-4 were projected respectively onto MoMac-VERSE. Germinal center, GC; interfollicular, IF; fold change, FC; macrophage signatures, MacroSigs.