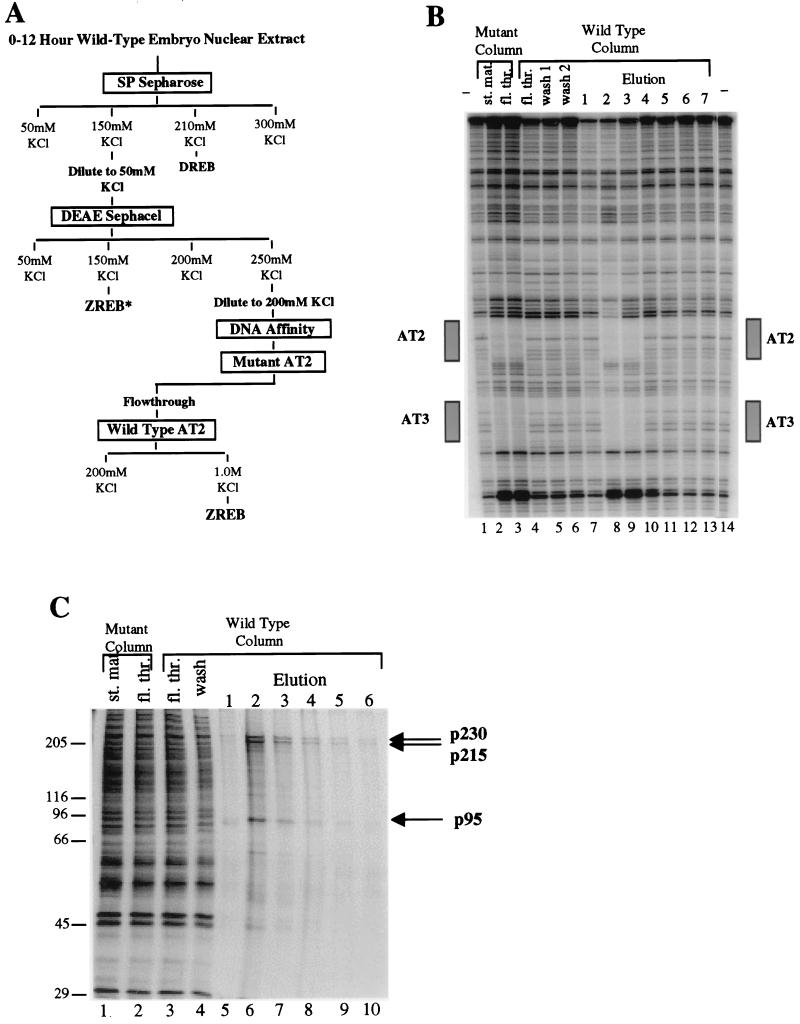
FIG. 1.
Purification of ZREB. (A) Flow chart of the ZREB purification scheme. Columns were eluted in a stepwise fashion with the indicated concentrations of KCl. DREB is the decapentaplegic repression element binding protein, which is equivalent to NTF-1 (18). Column fractions were assayed for AT2 site-specific DNase I footprinting activity. ZREB activity was purified to near homogeneity from the DEAE Sephacel 0.25 M KCl step by multiple rounds of DNA affinity chromatography. Another peak of AT2 binding activity (termed ZREB*) was detected in the DEAE Sephacel 0.15 M KCl step. This was later found to represent a degraded form of ZREB. (B) DNase I footprinting of material from the last two steps in the purification of ZREB. Lanes 1 and 14, no protein controls; lanes 2 and 3, 25 μl of starting material and flowthrough from mutant AT2 site affinity column. The flowthrough from the mutant AT2 site column was applied to the wild-type AT2 column. Footprints of fractions from this column are shown in lanes 4 to 13. Lane 4, 25 μl of flowthrough; lanes 5 and 6, 12.5 μl of serial 0.25 M KCl washes; lanes 7 to 13, 2.5 μl of serial 1 M KCl washes. The boxes beside the gel designate the location of the AT2 and AT3 elements in the zen VRR probe. (C) Silver-stained SDS-PAGE gel of the affinity column fractions. Lanes 1 and 2 contain the mutant affinity column starting (st.) and flowthrough (fl. thr.) fractions (2.5 μl). Lanes 3 and 4, wild-type affinity column flowthrough and wash fractions; lanes 5 to 10, column fractions eluted with 1.0 M KCl (12 μl). Molecular size standards (in kilodaltons) are indicated to the left. The positions of the three major polypeptide species (p230, p215, and p95) coeluting with the footprinting activity are indicated by arrows.