FIG. 3.
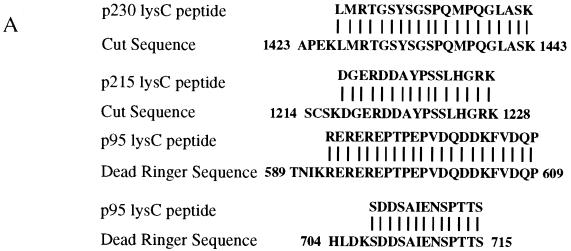
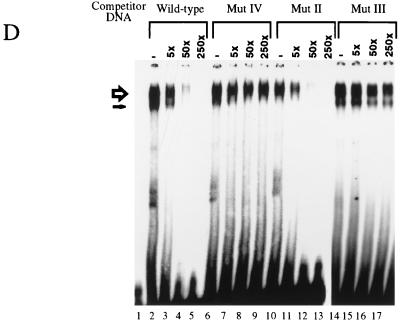
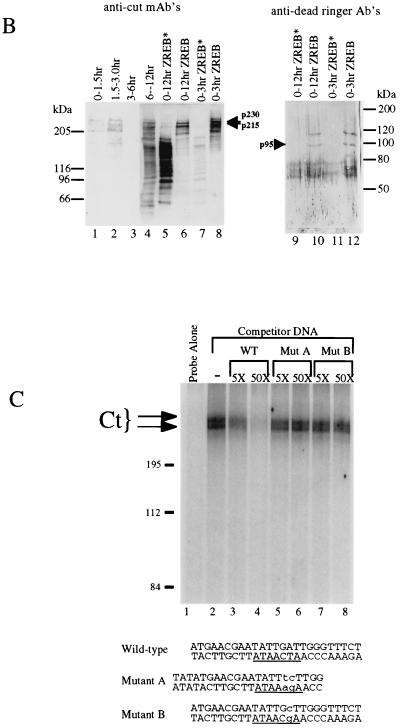
Identification of ZREB. (A) Approximately 10 pmol of p230, p215, and p95 were digested with endoproteinase Lys-C (13). Peptide fragments were purified by high-pressure liquid chromatography and sequenced by automated Edman degradation with a Perkin-Elmer/Applied Biosystems Division device (model 494), and the amino acid sequences were subjected to a BLAST sequence search. Results of the search are shown for each peptide. Both p230 and p215 peptides were exact matches to the homeodomain-containing protein Cut. Two separate peptides were sequenced from p95, and both were identical to Dri. One of the p95 peptides contains an internal lysine residue, clearly resulting from incomplete Lys-C digestion, since another p95 peptide was sequenced that was identical to this peptide, except that it terminated after the lysine residue (data not shown). (B) Western blots of crude embryo extracts and of purified ZREB and ZREB* were probed with anti-Ct (lanes 1 to 8) or anti-Dri (lanes 9 to 12) monoclonal antibodies (mAb’s) or antibodies (Ab’s). Lane 1, nuclear extracts of 0- to 1.5-h embryos; lane 2, nuclear extracts of 1.5- to 3-h embryos; lane 3, nuclear extracts of 3- to 6-h embryos; lane 4, nuclear extracts of 6- to 12-h embryos; lanes 5 and 9, ZREB* prepared from 0- to 12-h embryos; lanes 6 and 10, ZREB prepared from 0- to 12-h embryos; lanes 7 and 11, ZREB* prepared from 0- to 3-h embryos; lanes 8 and 12, ZREB prepared from 0- to 3-h embryos. In the anti-Dri immunoblot, the band above p95 with an apparent molecular size of about 115 kDa corresponds to a minor species seen in many ZREB preps and may represent full-length Dri, while p95 probably represents a truncated form of Dri. The series of bands extending downward from the 80-kDa marker represent a keratin contaminant in the ZREB and ZREB* preps against which the anti-Dri antibodies avidly react. (C) DNA affinity-purified ZREB was cross-linked to an iodinated DNA probe. The sequence of the probe is the same as that of the wild-type competitor, except that thymine residues were replaced with 5-iodouracil. All fractions were exposed to UV treatment. Lane 1, probe alone without protein; lanes 2 to 8, probe and 5 μl of affinity-purified ZREB; lanes 3 and 4, 5- and 50-fold excesses of cold wild-type (WT) competitor, respectively; lanes 5 and 6, 5- and 50-fold excesses of cold mutant competitor A, respectively; lanes 7 and 8, 5- and 50-fold excesses of cold mutant competitor B, respectively. Sequences of the wild-type and mutant competitors are indicated below the gel. Lowercase letters indicate mutations. (D) Recombinant Dri was assayed for binding to the AT2 site by electrophoretic mobility shift assays. The sequence of one strand of the radiolabeled double-stranded probe is TATGAACGAATATTGATTGGGA (the 7-bp core of the AT2 site is underlined). Lane 1, radiolabeled probe alone (100 fmol); lanes 2 to 17, recombinant Dri (250 ng) and radiolabeled probe (100 fmol). In addition to the radiolabeled probe and Dri, the binding reactions included 5-fold (lanes 3, 7, 11, and 15), 50-fold (lanes 4, 8, 12, and 16), or 250-fold (lanes 5, 9, 13, and 17) molar excess of the indicated cold competitors. The AT2-WT competitor is identical in sequence to the labeled probe. The AT2-mII, AT2-mIII, and AT2-mIV competitors have the same mutations as mutants II, III, and IV, respectively, in Fig. 2.