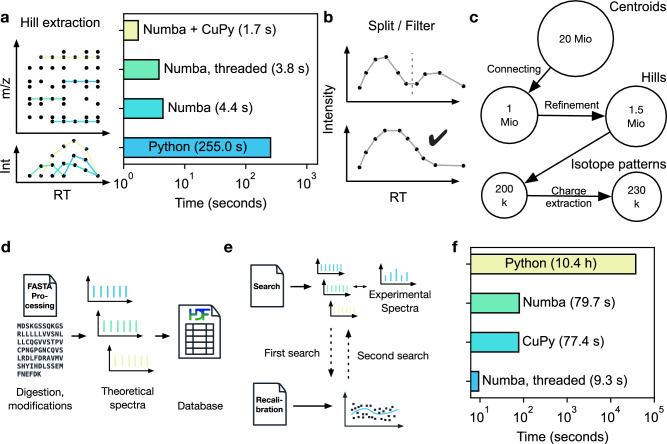
Fig. 3. Extracting isotope features and database search.
a Individual MS peaks of similar masses are connected over the retention time using a graph approach, resulting in “hills”. Using a native Python implementation, hill extraction takes several minutes. Numba, parallelization on CPUs or GPUs reduces hill extraction to seconds. CuPy refers to using GPU, Numba for single-threaded implementation, and Numba threaded to Numba using multiple threads. b Extracted hills are refined by splitting at local minima and only allowing well-formed elution profiles. c Starting with 20 million points for a typical Thermo HeLa shotgun proteomics file, these are connected to approximately one million hills, which increased to 1.5 million after hill splitting and filtering. Subsequent processing results in 200,000 pre-isotope patterns that ultimately yield 230,000 isotope patterns due to assignment to specific charge states. d The FASTA processing notebook contains functionality to calculate fragment masses from FASTA files which are saved in an HDF5 container for subsequent searches. e Initially, a first search is performed, and masses are subsequently recalibrated. Based on this recalibration, a second search with more stringent boundaries is performed. f Using the decorator strategy, the search can be drastically sped up, from 10 h in a pure Python implementation to seconds with Numba and CuPy.