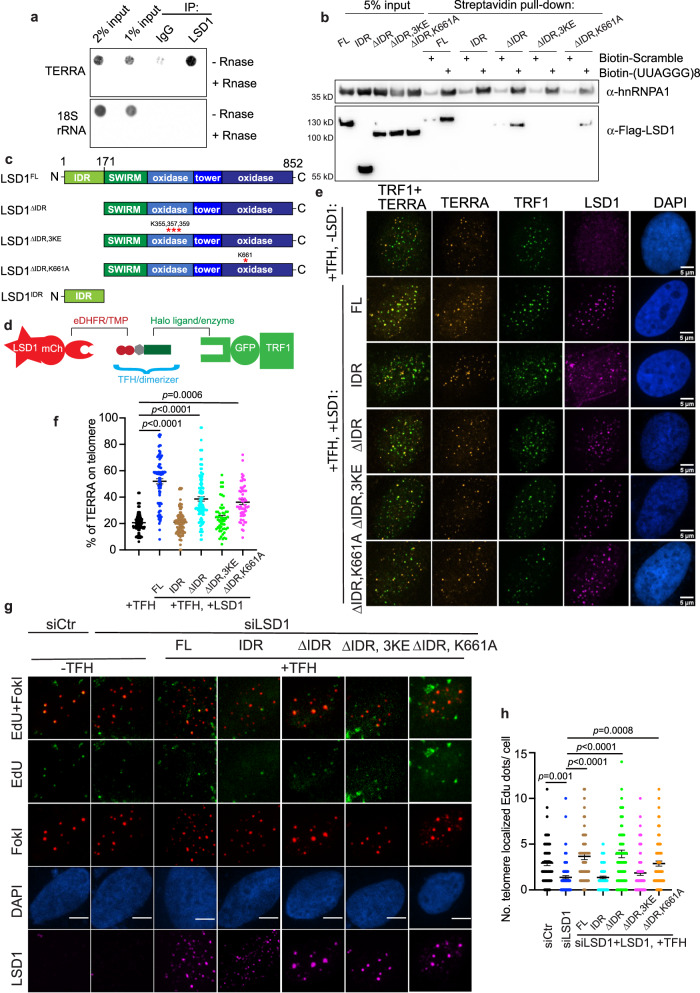
Fig. 3. Chemical dimerization of LSD1 to telomeres is sufficient to enrich TERRA and rescue ALT phenotypes.
a IP-recovered RNA was detected via dot-blot with probes annealing with TERRA or 18S rRNA (refer to Supplementary Fig. 4a for IP efficiency). b RNA pull-down assay was used to evaluate the binding of Streptavidin bead-bound biotinylated (UUAGGG)8 oligonucleotide to flag-tagged LSD1FL, LSD1IDR, LSD1ΔIDR, LSD1ΔIDR, 3KE, and LSD1ΔIDR, K661A from U2OS cell lysates (n = three independent experiments). c Schematic of LSD1: full-length (LSD1FL), C-terminus (LSD1ΔIDR,), N-terminus (LSD1IDR), mutant for nucleic acid binding (LSD1ΔIDR, 3KE), mutant for demethylation activity (LSD1ΔIDR, K661A). d Dimerization schematic: LSD1 is fused to mCherry and eDHFR, and TRF1 is fused to Halo enzyme and GFP. The dimerizer is TFH, TMP(trimethoprim)- Fluorobenzamide-Halo ligand. e Representative images and f quantification of TERRA (RNA FISH) localization on telomeres after dimerizing overexpressed LSD1FL, LSD1IDR, LSD1ΔIDR, and LSD1ΔIDR, 3KE, and LSD1ΔIDR, K661A to Halo-TRF1 in U2OS cells with dimerizer TFH (n = 108 cells for +TFH alone, n = 87 cells for LSD1FL, n = 103 cells for LSD1IDR, n = 113 cells for LSD1ΔIDR, n = 55 cells for LSD1ΔIDR, 3KE, n = 53 cells for LSD1ΔIDR, K661A over three independent experiments, one-way ANOVA). g Representative images and h quantification of Edu assay showing the rescue effect of newly synthesized telomeric DNA after dimerizing LSD1FL, LSD1IDR, LSD1ΔIDR, LSD1ΔIDR, 3KE, and LSD1ΔIDR, K661A to telomeres in siLSD1 cells (n = 69 cells for siCtr, n = 89 cells for siLSD1, n = 67 cells for LSD1FL, n = 77 cells for LSD1IDR, n = 90 cells for LSD1ΔIDR, n = 75 cells for LSD1ΔIDR, 3KE, n = 81 cells for LSD1ΔIDR, K661A over two independent experiments, one-way ANOVA). Error bars are mean ± SEM. NS not significant. Source data are provided as a Source Data file.