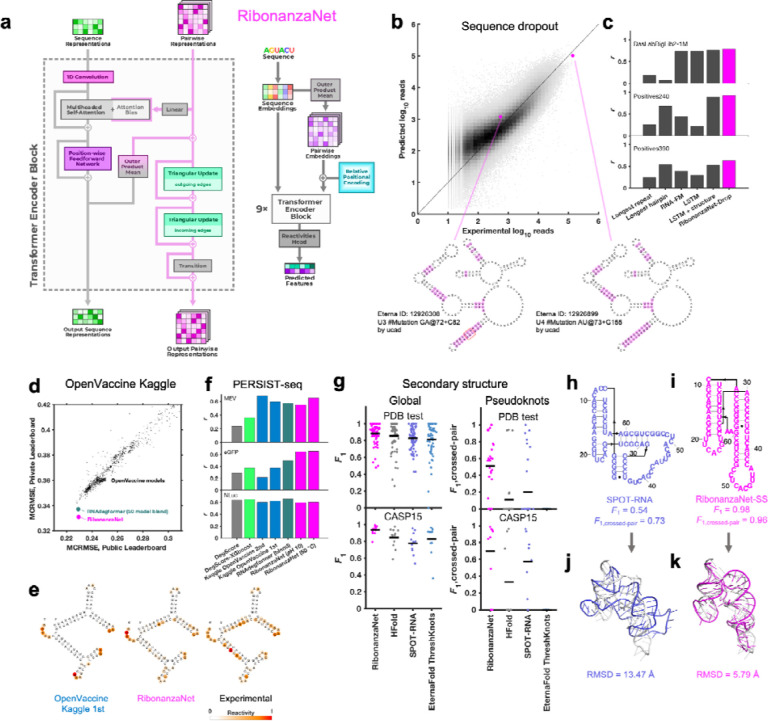
Figure 4. RibonanzaNet model and fine-tuning for downstream tasks.
(a) RibonanzaNet architecture unifies features of RNAdegformer and top Kaggle models into a single, self-contained model. (b) Predictions of RibonanzaNet-Drop for sequence dropout during SHAPE chemical mapping experiments, tested on DasLabBigLib2–1M after fine-tuning on Illumina sequence read counts in DasLabBigLib-1M. Diagrams depict similar sequences (differences highlighted in magenta; red rounded rectangle in left-hand diagram show G-C pairs stabilizing long stem) with identical predicted secondary structures (pseudoknot not shown) but different levels of dropout. (c) Pearson correlation coefficients of logarithm of sequencer read counts compared to RibonanzaNet-Drop and baseline models for three test sets. (d) Modeling accuracy of RibonanzaNet-Deg for OpenVaccine test sets (Public & Private Leaderboard) after fine-tuning on OpenVaccine training examples. MCRMSE: mean column root mean squared error for SHAPE reactivity and two degradation conditions (pH 10 and 50 °C, 10 mM MgCl2, 1 day). (e) SHAPE reactivity of OpenVaccine test molecule ‘2204Sept042020’ predicted by top model in Kaggle OpenVaccine competition (‘Nullrecurrent’) and RibonanzaNet, compared to experimental profile. (f) Pearson correlation coefficients of degradation profiles predicted by different algorithms compared to degradation rates measured by PERSIST-seq for mRNA molecules encoding a multi-epitope vaccine SARS-CoV-2 (MEV), enhanced green fluorescent protein (eGFP), and nanoluciferase (NLuc). (g) Secondary structure accuracies of RibonanzaNet-SS and other packages on a temporally split PDB test set (top) and CASP15 RNA targets (bottom). F1 is harmonic mean of precision and recall of base pairs; lines in violin plots display mean F1. (h-i) Secondary structure models for CASP15 target R1107, human CPEB3 ribozyme, derived from (h) SPOT-RNA and (i) RibonanzaNet.38 (j-k) Overlay of X-ray structure (white) and 3D models using trRosettaRNA guided by secondary structures from (j) SPOT-RNA and (k) RibonanzaNet.