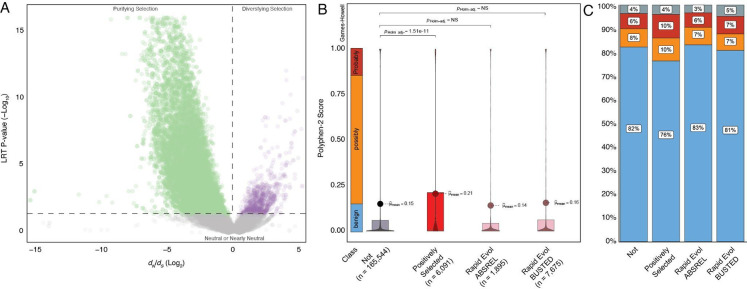
Figure 2. Amino acid substitutions in positively selected genes are likely deleterious.
A. Volcano plot showing the dN/dS (Log2) and likelihood ratio test (LRT) P-value (−Log10) for codons with elephant-specific amino acid changes in positively selected and rapidly evolving genes. dN/dS rates and P-values were estimated with the fixed effect likelihood (FEL) model, the P-value is for the LRT of dN/dS ≠ 1 at each codon. Codons with dN/dS>1 and a P-value≤0.05 are colored purple, those with N/dS<1 and a P-value≤0.05 are colored green, and those with dN/dS ≈ 1 are colored grey.
B. Violin and box plots showing the distribution of PolyPhen-2 scores for elephant-specific amino acid changes. The number of amino acid substitutions is given in parenthesis for each gene set. The range of scores corresponding to benign, possibly damaging, and probably damaging mutations is highlighted. Multiple hypotheses (Holm) corrected p values (pHolm-adj.) are shown for statistically significant comparisons; NS, not significant. Not, amino acid changes in genes with no evidence of positive selection or rapid evolution; Positively selected, amino acid changes in positively selected genes; Rapid Evol ABSREL, amino acid changes in rapidly evolving genes inferred from ABSREL; Rapid Evol BUSTED, amino acid changes in rapidly evolving genes inferred from BUSTED.
C. Bar chart showing the qualitative PolyPhen-2 predictions of amino acid changes. Not, amino acid changes in genes with no evidence of positive selection or rapid evolution; Positively selected, amino acid changes in positively selected genes; Rapid Evol ABSREL, amino acid changes in rapidly evolving genes inferred from ABSREL; Rapid Evol BUSTED, amino acid changes in rapidly evolving genes inferred from BUSTED.