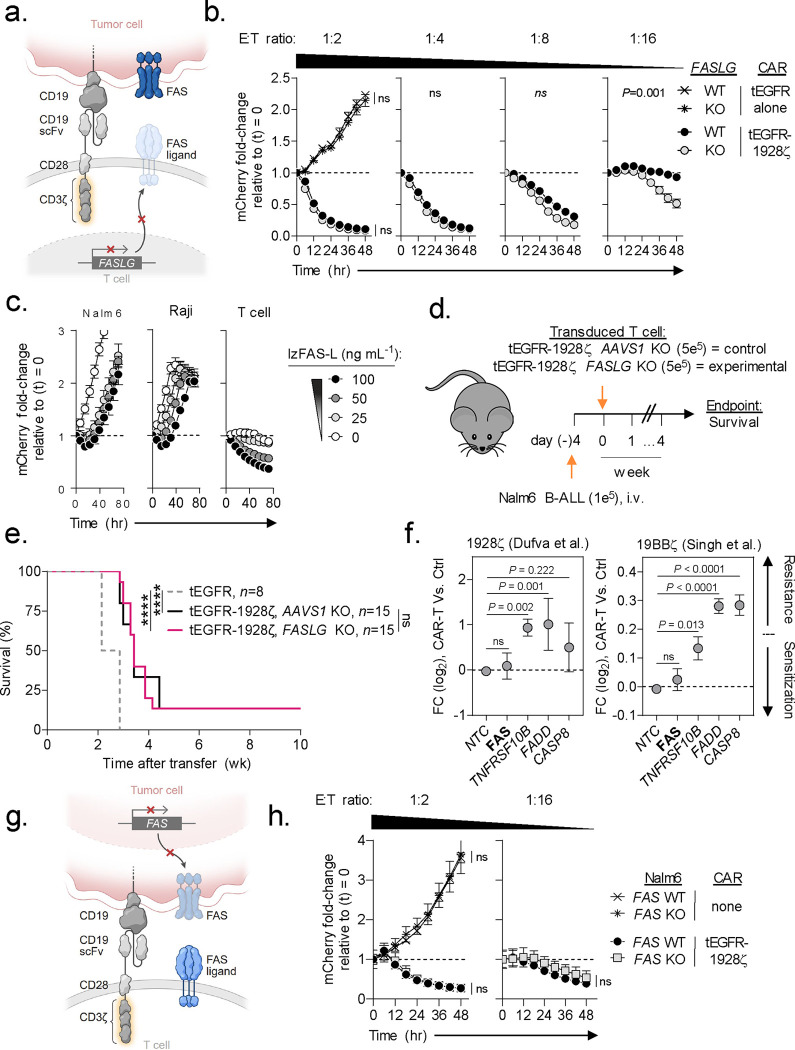
Fig. 4: FAS-L/FAS-signaling is dispensable for CAR-T antitumor efficacy.
(a) Schematic for the CRISPR/Cas9-mediated knockout (KO) of FASLG (experimental) or AAVS1 (control) in human CD8+ T cells expressing a 1928ζ CAR. (b) Cytolytic activity of FASLG-KO versus AAVS1-KO 1928ζ CAR-T cells against Nalm6/mCherry at indicated effector to target (E:T) ratios. Data shown as mean ± s.e.m. using n=3 biological replicates per condition. Statistical comparisons performed using a one-way ANOVA. ns = not significant (P>0.05). (c) Growth kinetics of Nalm-6 B-ALL, Raji B-NHL, or activated T cells in the presence or absence of lzFAS-L. Each cell type was transduced with mCherry. Data shown as mean ± s.e.m. using n=3 biological replicates per condition following incubation with an indicated concentration of lzFAS-L. hr, hour. (d) Experimental design and (e) Kaplan–Meier survival curve comparing the in vivo antitumor efficacy of human CD8+ T cells that express a 1928ζ CAR with FASLG-KO or AAVS1-KO against established Nalm-6 B-ALL. tEGFR alone, n=8; tEGFR-1928ζ AAVS1-KO, n=15; tEGFR-1928ζ FASLG-KO, n=15. ****P< 0.0001 and ns (not significant, P>0.05) using log-rank test. wk, week. (f) Scatter plots displaying the enrichment or depletion of synthetic guide RNAs (sgRNAs) targeting indicated genes in the death receptor pathway by Cas9-expressing Nalm-6 B-ALL cells. Tumor cells were placed under selection by T cells transduced with a 1928ζ CAR (left), a 41BBζ CAR (right), or left non-transduced as specificity controls (Ctrl). Data re-analyzed from two published genome-scale CRISPR/Cas9 screens56,57 and is shown as mean log2 fold-change (FC) ± s.e.m of sgRNAs targeting indicated genes. NTC = non-targeted control sgRNAs. Gene level significance was determined using a one-way ANOVA corrected for multiple comparisons by Dunnett’s test. (g) Schematic for the CRISPR/Cas9-mediated knockout (KO) of FAS using an individual sgRNA in Nalm6 B-ALL. (h) Time-dependent cytolytic activity of 1928ζ CAR-T cells against FAS-KO versus FAS-wild type (WT) Nalm6/mCherry cells at a high (left) or low (right) E:T ratio. Data shown as mean ± s.e.m. using n=3 biological replicates per condition and time point. Statistical comparisons were performed using a one-way ANOVA.