Figure 4. ORF48 deletion defects KSHV lytic replication.
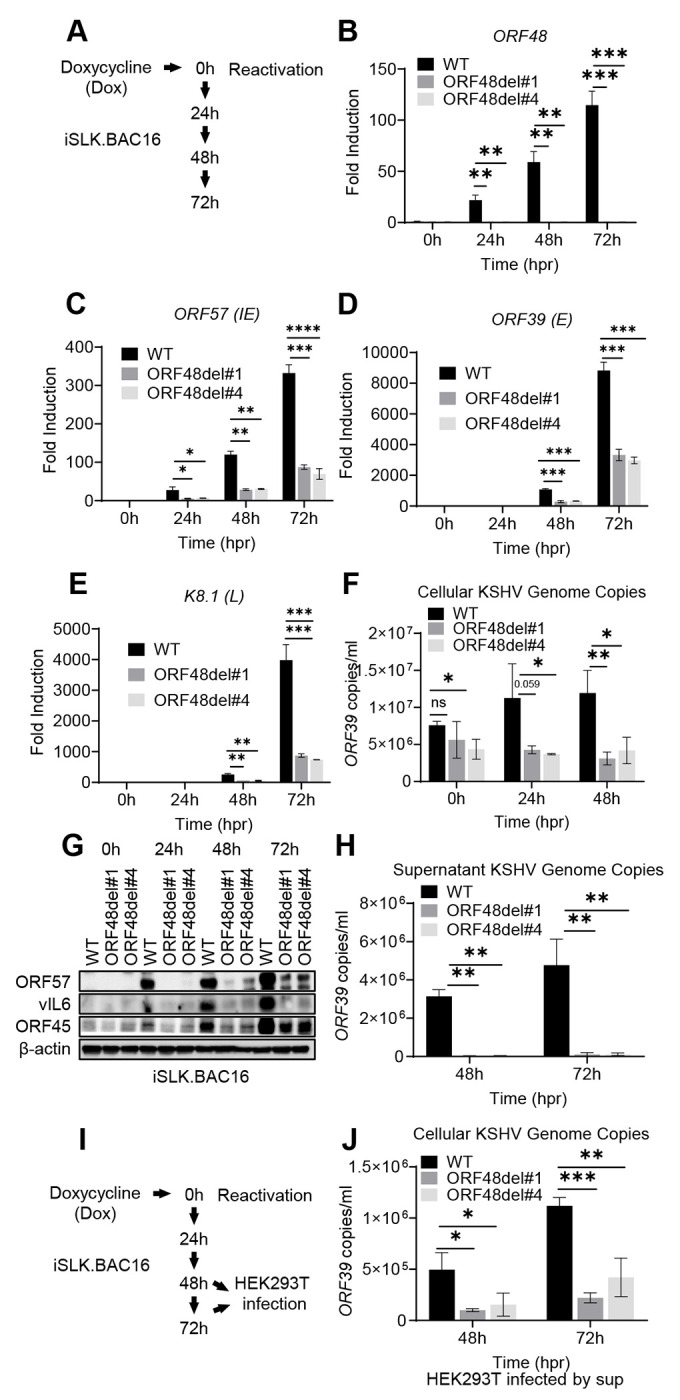
(A) Schematic illustration of the experimental procedure of (B-H). Total RNAs were extracted from all groups at all time points to synthesize cDNA and subjected to RT-PCR. Specific RT-PCR primers were used to detect (B) ORF48 for deletion confirmation, (C) ORF57 representing an immediate early lytic gene, (D) ORF39 representing an early lytic gene, and (E) K8.1 representing a late lytic gene. Expression levels of these genes were normalized with GAPDH. (F) Cellular KSHV genome copies were quantitated using a genomic primer based on the ORF39 coding sequence as previously described. A STREP-tagged ORF39 (37) was used to generate the standard curve. (G) Western blot analysis of ORF57, vIL6 and ORF45 encoded by KSHV. (H) The supernatants from all 48h and 72h groups containing KSHV genome copies were quantitated using the same method as (F). (I) Schematic illustration of the experimental procedure for infection assay. Briefly, the supernatants from all 48h and 72h groups were collected to infect naïve HEK293 cells to evaluate infectious virion production. (J) Forty-eight hours post-infection, genome DNAs from infected HEK293 cells were extracted and the KSHV genome copy numbers were evaluated by the same method as (F). Data are presented as mean ± s.d. from at least three independent experiments. *indicates p<0.05. ** indicates p<0.01 *** indicates p<0.001 **** indicates p<0.0001 by Student’s t-test.
