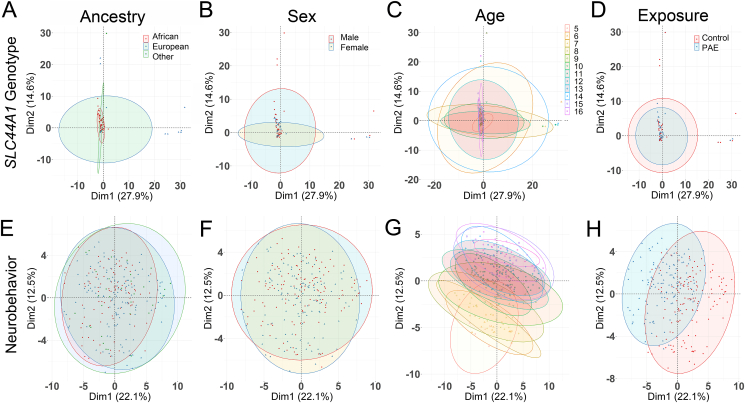
FIGURE 2.
Principal component analysis of SLC44A1 genotypes and neurobehaviors against population characteristics. (A–D) Principal component analysis (PCA; FactoMineR, v2.3) was used to examine the genomic separation of all participants based on ancestry, sex, age, and exposure status. (A) Genetic ancestry based on comparison against 1000 Genomes vs. genotype. Red, African; Blue, European; Green, Other. (B) Sex vs. genotype. Red, males. Blue, females. (C) Age vs. genotype, stratified in 1-y intervals. (D) Exposure status vs. genotype. Red, low-to-no prenatal exposure (PAE); Blue, heavy PAE. No significant associations between genotype and these population characteristics were observed in the PCA. (E–H) PCA to examine the separation of neurobehaviors in all participants based on ancestry, sex, age, and exposure status. All color symbols as described for (A–D). (E) Genetic ancestry vs. neurobehavior. (F) Sex vs. neurobehavior. (G) Age vs. neurobehavior. (H) Exposure status vs. neurobehavior. There was no influence of ancestry and sex on neurobehavior. PAE status contributed to the behavioral variance in PC1. Age contributed to the behavioral variance in PC2. The 95% confidence interval surrounding each group mean is indicated by the ellipseing.