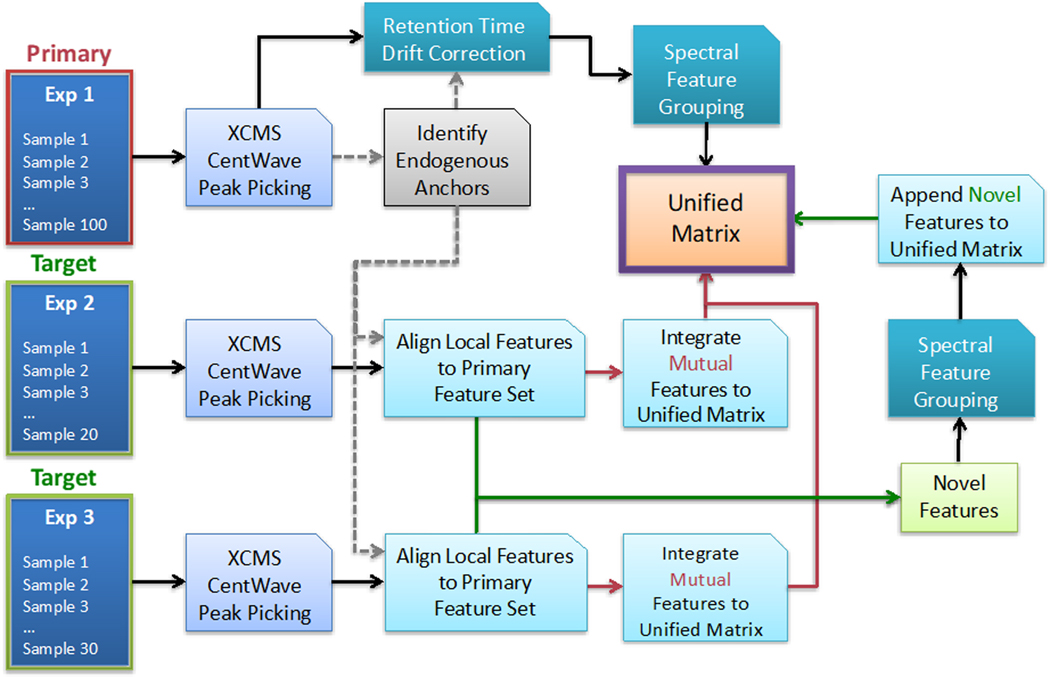
Figure 1.
An overview of the Disparate Metabolomics Data Reassembler (DIMEDR) workflow for integrating potentially incongruent datasets originating from multiple experiments. The algorithm relies on an initial selection of a “primary” dataset, with all other datasets designated as “target” sets. The primary dataset can be the largest or, what is determined to be the highest quality, and serves as the template for creating the unified matrix that data from all other target sets will be integrated into. This is facilitated by extracting persistent spectral features from the primary dataset, called endogenous anchors, that are utilized as reference points for universal data correction. Endogenous anchors are initially identified in each target dataset, and subsequently used to align, identify, and integrate mutual spectral features that are shared with the primary dataset into the unified matrix. Novel features, for which retention times have also been corrected via this process, are collected across all target datasets, grouped, and appended to the unified matrix as well.