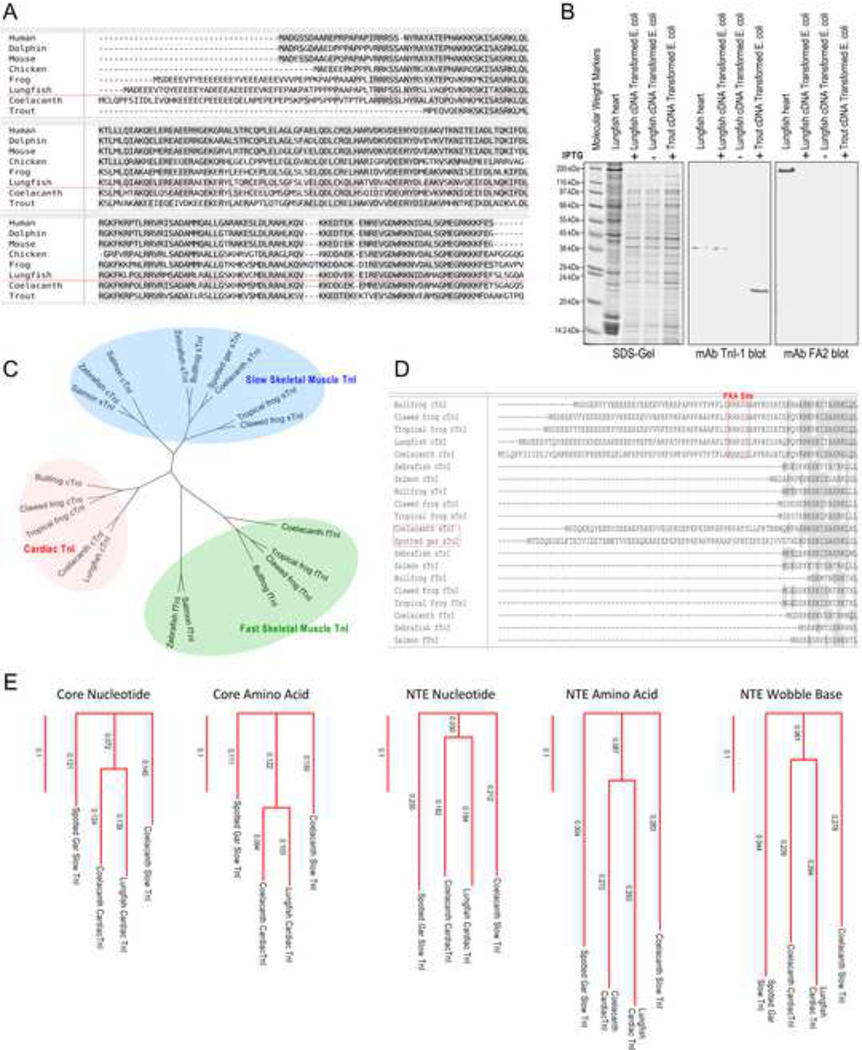
Fig. 6. The primary structure of lungfish cardiac TnI and the presence of N-terminal extension containing the two PKA substrate serines.
A, The amino acid sequence of lungfish cardiac TnI is aligned with cardiac TnI from representative vertebrate species. The result shows that lungfish cardiac TnI has a conserved core structure similar to cardiac TnI in the other species. In contrast to cardiac TnI of ray-finned fish, lungfish cardiac TnI has a long N-terminal extension similar to that of coelacanth and amphibian cardiac TnIs. Despite of the significant difference in length, the N-terminal extension of lungfish, coelacanth, amphibian, avian and mammalian cardiac TnIs all have the two PKA substrate serine residues and similar flanking sequences. B, The SDS-gel and mAb TnI-1 Western blot show that the lungfish cardiac TnI protein expressed in E. coli from the cloned cDNA has a gel mobility same as that of native cardiac TnI in the heart of spotted African lungfish (Protopterus dolloi). In comparison, E. coli expressed rainbow trout cardiac TnI that lacks the N-terminal extension (GenBank accession # HM012798.1) showed a significantly faster mobility. The Western blot using mAb FA2 against cardiac MHC further confirmed the mobility position of MHC band in our SDS-PAGE conditions. C, The phylogenetic tree shows that lungfish cardiac TnI is closely related to coelacanth and amphibian cardiac TnIs. In contrast, cardiac and slow skeletal muscle TnIs in ray-finned fishes are not specifically diverged and both lack the N-terminal extension while fish fast TnI is closely related to amphibians. D, Alignment of N-terminal segment of cardiac TnI from multiple fish and amphibian species revealed that besides cardiac TnI, slow skeletal muscle TnI of coelacanth and spotted gar also have a long N-terminal extension but lacking the two PKA phosphorylated serine residues, suggesting a possible transitional state. E, The phylogenetic trees showed that the N-terminal extensions (NTE) found in fish TnIs are less conserved than the core structures and the degree of divergence is higher at amino acid level than that at nucleotide level, indicating a low degree of functional convergence. A tree constructed using the wobble bases of N-terminal extension codons is shown as a reference level of low functional selection.