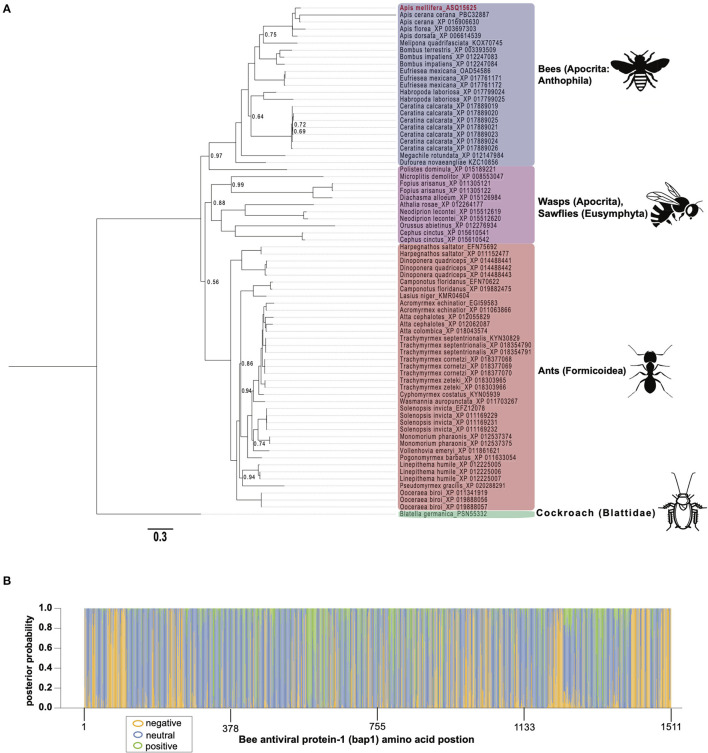
Figure 2.
Bee antiviral protein-1 phylogenic relationship to other Hymenopteran orthologs inferred from amino acid sequence. (A) Majority rule Bayesian consensus tree of Bap1 homologs derived from Bayesian analysis of amino acid alignment implemented in Mr. Bayes v3.2 using a Jones substitution model. Numbers on branches and nodes are posterior probabilities (0-1), though posterior probabilities values of 1 are not shown to improve clarity. The scale bar corresponds to proportion of amino acid change. Accession numbers are included on the branch tips and in Supplementary Table S2. (B) A corresponding majority Rule Bayesian consensus tree derived from Bayesian analysis of a codon alignment was used in a selection analysis in the CODEML package in Phylogenetic Analysis by Maximum Likelihood (PAML) 4.9. The Bayes Empirical Bayes method under model 2, which assumes 3 site classes (negative, neutral, and positive selection), was used to calculate the posterior probability that each amino acid position belonged to each site class. The posterior probabilities were then plotted as a stacked bar chart along the length of Bap1 amino acids. This method shows clear diffuse neutral selection with regions under strong positive or negative selection.