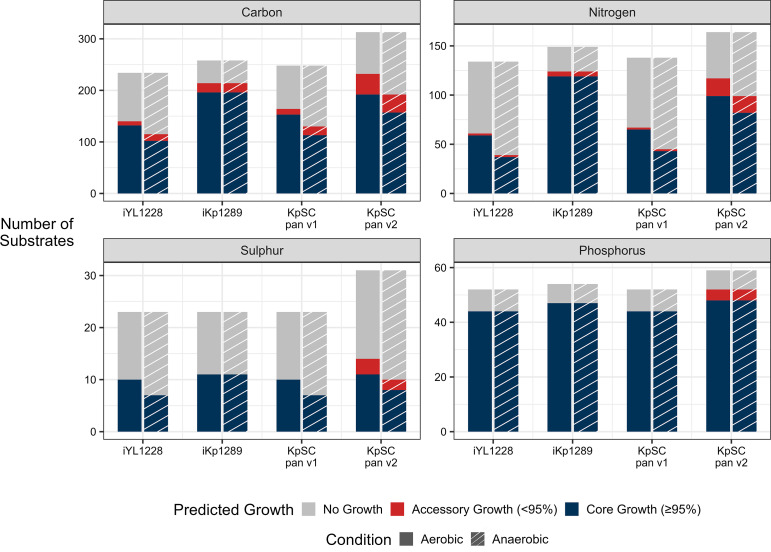
Fig. 3.
Growth simulation outcomes for strain-specific genome-scale metabolic models derived from four different references. Simulations represent growth in minimal media supplemented with a single source of carbon, nitrogen, sulphur or phosphorus as indicated. The total number and set of substrates simulated is dependent upon those that can be supported by the reference model (see Table S4; note that substrates supported by the iYL1228 reference are a subset of those supported by KpSC pan v1, which are a subset of those supported by KpSC pan v2. Substrates supported by iKp1289 are an independent and overlapping set). Core growth (blue) refers to substrates for which growth was predicted for ≥95 % of the population, whereas accessory growth (red) refers to substrates for which growth was predicted for <95 % of the population. ‘No growth’ (grey) refers to metabolites that are supported by the model, but do not result in positive growth predictions for any strains. These can occur due to incomplete metabolic pathways in the model (e.g. the carbon utilization pathway is complete but the nitrogen pathway is not, resulting in no growth on nitrogen) or where Klebsiella is legitimately unable to utilize the substrate for growth.