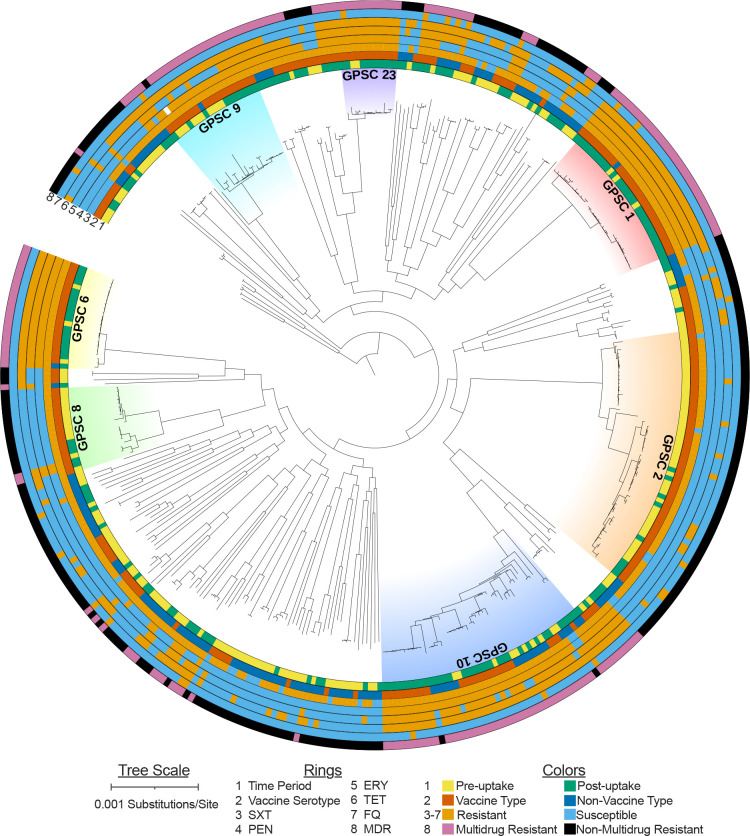
Fig. 2.
Population structure of invasive pneumococci in southern India. Maximum-likelihood tree, with branch lengths corrected for recombination events, is based on a quality-filtered core genome alignment with invariant nucleotides and biallelic SNPs. Seven prevalent GPSCs (global pneumococcal strain clusters or strain lineages) are indicated. The rings indicate isolate characteristics analysed in the text including: isolation date in the PCV pre- or post-uptake time periods as defined according to Fig. 1, vaccine type or non-vaccine type serotype defined by coverage in PCV13-Prevnar, and antimicrobial resistance determinants.