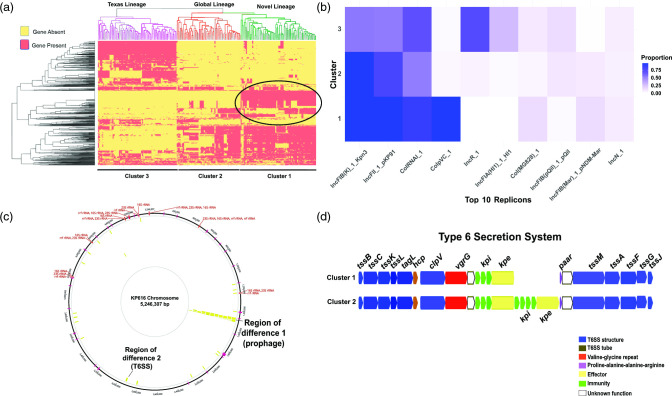
Fig. 6.
Assessment of accessory genome differences between CG307 strains. (a) Gene presence/absence heatmap representing the agglomerative hierarchical clustering of accessory genomes of CG307 strains. The circled region indicates genetic content unique to cluster 1 isolates. (b) Proportion heatmap of the most common replicon types identified using PlasmidFinder stratified by CG307 cluster. (c) Recombination hotspots (yellow blocks) of CG307 isolates using Kp616 as a reference and Gubbins predicted recombination sites output. Regions labelled on the chromosome indicate genomic context we identified that differed between cluster 1 and 2. (d) Region of difference 2 between CG307 cluster 1 and cluster 2 strains involving a T6SS. The labelling system for T6SS proteins is taken from another study [76] with a description of the colours used for putative gene functions shown in the key. Note that cluster 1 strains lack of a set of immunity/effector proteins present in cluster 2 strains.