Fig. 5. Analysis of gut microbiome profile and the relative abundance of phyla and class level difference after polymicrobial infection as measured in the feces of APP/PS1 mice.
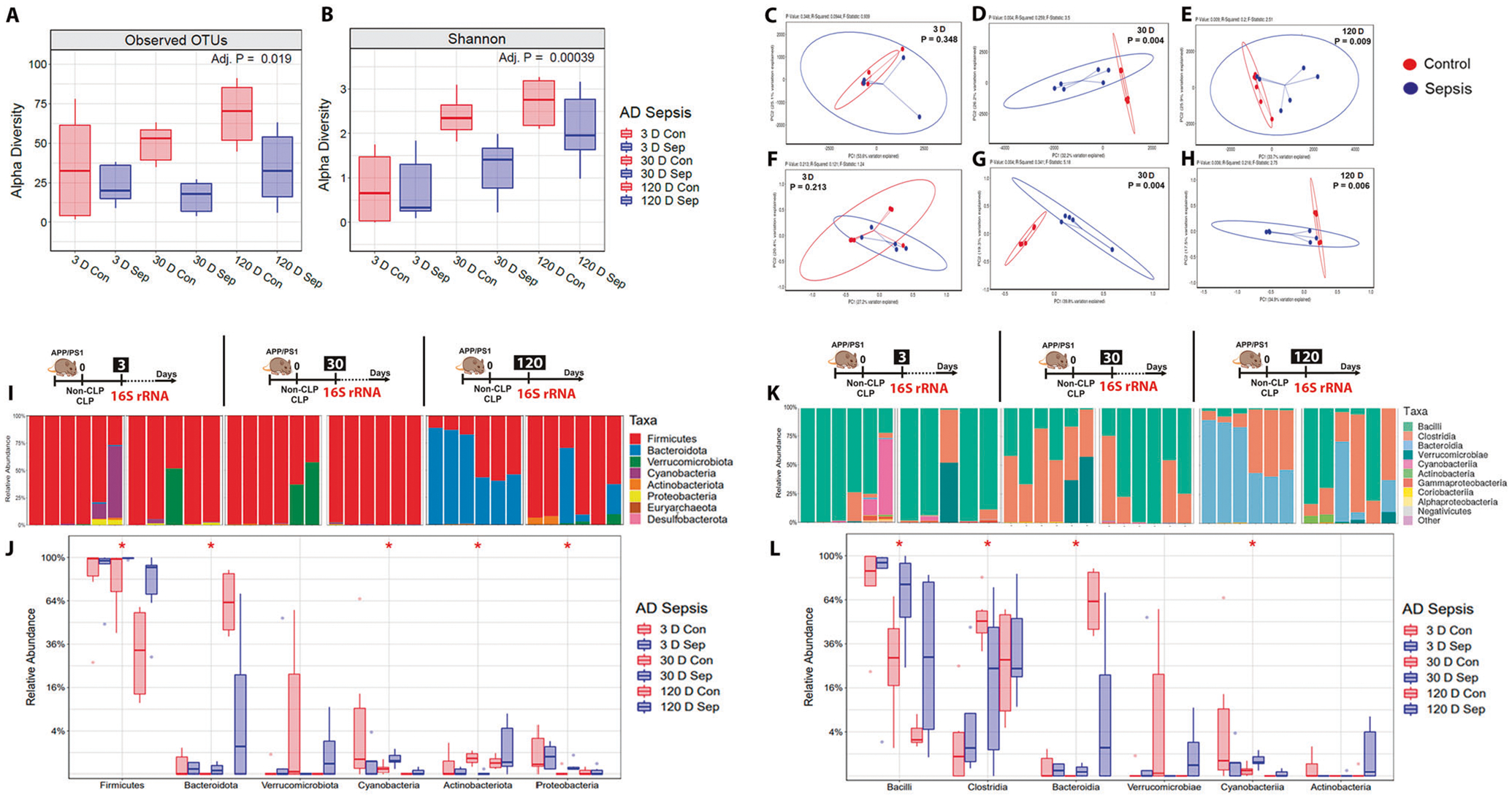
Using 16S rRNA sequencing, the α-diversity (microbiota richness) and β-diversity (difference in global microbiota composition between samples) were determined at 3, 30, and 120-time points after CLP and non-CLP surgery. A α-diversity: Observed Operational Taxonomic Units (OTUs). B α-diversity: Shannon-diversity index. At 30 and 120 days after sepsis induction, there was a significant reduction in the α-diversity difference in sepsis-surviving mice compared to the control group. C and F β-diversity at three days: Principal coordinate analysis using unweighted and weighed UniFrac. D and G β-diversity at 30 days: Principal coordinate analysis using unweighted and weighted UniFrac, p = 0.004. E and H β-diversity at 120 days: Principal coordinate analysis using unweighted UniFrac, p = 0.009 and weighted UniFrac, p = 0.006, n = 6. The 16S rRNA sequencing was used to measure the relative abundance of the phyla and class at 3, 30, and 120-time points after CLP and non-CLP surgery. I Relative abundance stacked bar plots of taxa from CLP or non-CLP-treated mice at the phylum level. J Quantification of taxa at the phylum level. No change was found at 3 and 120 days, p ≥ 0.05. At 30 days, the phylum Actinobacteria decreased, and the phylum Bacteriodetes, Cyanobacteria, Firmicutes, and Proteobacteria increased in the sepsis group compared with the control group, p < 0.05. K Relative abundance stacked bar plots of taxa from at the class level. L Quantification of taxa at the class level. Similarly, there was no change at the class level in 3 and 120 days, p ≥ 0.05). The class Bacilli, Bacteroidia, and Cyanobacteria increased, but the class Clostridia decreased in the sepsis group at the intermediate time point, p < 0.05, n = 6. For 16S rRNA sequencing, Kruskal–Wallis test with post hoc Benjamini-Hochberg correction was used for statistical analysis.
