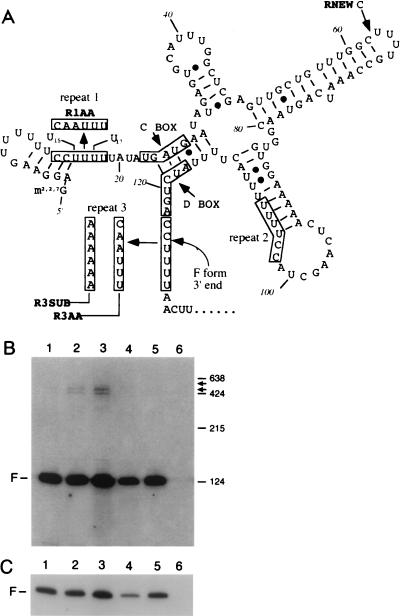
FIG. 9.
Effects of mutations in snR13 on RNA accumulation. (A) Predicted secondary structure of wild-type snR13F RNA, generated by using the Genetics Computer Group Fold program (11). The locations of the C and D boxes, the primer extension stops, and the mutations R1AA, R3SUB, R3AA, and RNEW are shown (see text). The 3′ end of wild-type snR13F is formed between residues C124 and U125. (B) Effects of snR13 mutations. Strain TR110, which carries snR13-Δ1, was transformed with plasmids as indicated below. Transformants were grown at 30°C followed by RNA extraction and Northern blotting using oligonucleotide 13A (Fig. 1) as the probe. Lane 1, pTR48 (wild-type SNR13); lane 2, pTR51 (snR13-R3AA); lane 3, pTR53 (snR13-R3SUB); lane 4, pTR52 (snR13-R1AA); lane 5, pTR56 (snR13-RNEW); lane 6, pRS315 (vector only). Arrows to the right indicate the mobilities of the novel RNAs. Size standards are indicated in nucleotides to the right. (C) A shorter exposure of the Northern blot shown in panel B shows that snR13F is decreased in abundance in lane 4 (see text).