Abstract
Lindera obtusiloba Blume is the northernmost tree species in the family Lauraceae, and it is a key species in understanding the evolutionary history of this family. The species of L. obtusiloba in East Asia has diverged into the Northern and Southern populations, which are geographically separated by an arid belt. Though the morphological differences between populations have been observed and well documented, intraspecific variations at the plastomic level have not been systematically investigated to date. Here, ten chloroplast genomes of L. obtusiloba individuals were sequenced and analyzed along with three publicly available plastomes. Comparative plastomic analysis suggests that both the Northern and the Southern populations share similar overall structure, gene order, and GC content in their plastomes although the size of the plasome and the level of intraspecific variability do vary between the two populations. The Northern have relatively larger plastomes while the Southern population possesses higher intraspecific variability, which could be attributed to the complexity of the geological environments in the South. Phylogenomic analyses also support the split of the Northern and Southern clades among L. obtusiloba individuals. However, there is no obvious species boundary between var. obtusiloba and var. heterophylla in the Southern population, indicating that gene flow could still occur between these two varieties, and this could be used as a good example of reticulate evolution. It is also found that a few photosynthesis‐related genes are under positive selection, which is mainly related to the geological and environmental differences between the Northern and the Southern regions. Our results provide a reference for phylogenetic analysis within species and suggest that phylogenomic analyses with a sufficient number of nuclear and chloroplast genomic target loci from widely distributed individuals could provide a deeper understanding of the population evolution of the widespread species.
Keywords: complete chloroplast genomes, intraspecific diversity, Lindera obtusiloba, phylogenetic analysis, positive selection
Lindera obtusiloba in East Asia has diverged into the Northern and Southern populations, which are geographically separated by an arid belt. Here, ten chloroplast genomes of L. obtusiloba species were sequenced and analyzed along with three publicly available plastomes support the split of the Northern and the Southern clades and found a fewer photosynthesis‐related genes were under positive selection, which mainly related to the difference geological environments between Northern and Southern regions.

1. INTRODUCTION
Intraspecific variation is defined as genotypic and phenotypic diversity observed within a given species. Despite its significance in the taxonomic division, ecological processes, food security, and medicinal usage (Des Roches et al., 2018; Wilting et al., 2015), intraspecific variation has barely been taken into account when assessing biodiversity in current days (Díaz et al., 2018; Mimura et al., 2017; Ruckelshaus et al., 2020). Most evolutionary studies focus on interspecific variations (Alix et al., 2017; Aschehoug et al., 2016; Martín‐Serra & Benson, 2020) or genetic differentiation within a population (Halbritter et al., 2018; Taudt et al., 2016). Some efforts have been put into understanding the influence of biogeographic boundaries, environmental variables, and historical climatic instability on population genetics and phenotypic structure, especially intraspecific variations in species with a wide geographical distribution (Gray et al., 2019; Huang et al., 2016; Ye et al., 2017; Zhang et al., 2016). The demographic history of some species proves that subspecies living in separated subregions have experienced different selection pressures and have accumulated various mutations in their genomes (Tian et al., 2020). Intraspecific variation was recognized as a prerequisite for speciation (Bürger et al., 2006). Thus, a better description of the complexity of the genomic divergence among intraspecies, as well as the evolutionary processes, might help us identify species at risk of extinction and allow us to apply conservation strategies to protect them (Blomqvist et al., 2010; Leigh et al., 2019; Mimura et al., 2017).
Lindera Thunb. (spicebush or spicewood), one of the largest genera in the family Lauraceae, comprises almost 100 species, which are widely distributed in East Asia and East America (Chanderbali et al., 2001). Lindera is key to studying the origin and maintenance of tropical and subtropical plant communities, which are of significant economic and scientific value (Cao et al., 2016; Ye et al., 2017). Based on morphological characteristics (leaf venation, brachyblasts, pedunculate, fruit cupules, and so on), the genus Lindera has been divided into eight sections, namely sect. Aperula, sect. Cupuliformes, sect. Daphnidium, sect. Lindera, sect. Palminerviae, sect. Polyadenia, sect. Sphaerocarpae, and sect. Uniumbellae (Tsui, 1987). Due to the parallel evolution occurring between Lindera and Litsea Lam., proper classification of the two species or sections from the two genera proves to be difficult (Li, 1985). Previous studies have shown that nuclear loci (ITS, rpb2, etc.) and chloroplast regions (matK, trnK, trnL‐trnF, etc.) could not provide enough information to resolve the relationship between Lindera and Litsea and that species from the eight section of Lindera were placed in Litsea (Fijridiyanto & Murakami, 2009; Li, Conran, et al., 2008; Li, Li, & Li, 2008; Rohwer, 2000). Recently, some monophyletic clades, such as the sect. Aperula, sect. Polyadenia and trinerved Lindera complex have been successfully detected and classified via whole plastomic analyses (Fijridiyanto & Murakami, 2009; Li et al., 2004, 2007; Liu, Chen, et al., 2021; Liu, Ma, et al., 2021; Tian et al., 2019; Xiao et al., 2020). Although these detected monophyletic clades align well with morphology, the generic delimitation between Lindera and Litsea remains to be solved. Moreover, most phylogenetic studies have only sampled a single individual for each species, which is not sufficient for providing a consistent and reliable conclusion about the taxonomic status of that species (Liu, Chen, et al., 2021; Liu, Ma, et al., 2021; Xiao et al., 2022). It is also unclear whether the factors, such as hybridization or gene flow among intraspecific species, would drive taxonomic confusion among various varieties.
Lindera obtusiloba Blume is a deciduous shrub of the section Palminerviae in the genus Lindera, and it is widely distributed in the subtropical and temperate zones of East Asia. This species has diverged into var. obtusiloba and var. heterophylla, and this differentiation is based on the leaf margin morphology (Li, Conran, et al., 2008; Li, Li, & Li, 2008). The geological distribution of var. heterophylla is limited to the Southwest of East Asia while var. obtusiloba is widely distributed in East Asia. Despite of distributional difference, it is difficult to distinguish the two varieties based on morphology and molecular phylogeny in the Southern populations. It is of important economic value for it can be used as the raw material for the production of lubricants and fertilizers (Zekun & Haixia, 2012). Moreover, L. obtusiloba has also been used as a traditional medicinal herb for the treatment of liver damage and inflammation (Freise et al., 2010; Hong et al., 2013). L. obtusiloba is the Northernmost species in Lauraceae, and it is of great significance in studying the adaptation of Lauraceae plants to cold environments (Tian et al., 2020; Zhang et al., 2014). In previous studies, the divergence of L. obtusiloba into the Northern and Southern populations has been observed in studies with both chloroplast DNA and nuclear microsatellite markers (Ye et al., 2017). The two populations are geographically separated by an arid belt, which is modulated to be drier and wider during the Palaeogene (Guo et al., 2008). The arid belt acts as a barrier, which impedes gene flow between individuals living in the two sub‐regions (Bai et al., 2016; Chen & Lou, 2019; Ye et al., 2017), eventually leading to the formation of two separate populations (Tian et al., 2020; Xu et al., 2021). Previous studies have focused on population genetic diversity and the evolutionary history of this species. A divergence between the northern and southern populations of L. obtusiloba in East Asia has been observed in phylogenetic studies using cp DNA and SSRs. However, this divergence still lacks support from evidence at the genomic level.
The chloroplast is the organelle in which photosynthesis takes place, and it plays a vital role in energy conversion in plants (Ruhlman & Jansen, 2014). Chloroplast is a semi‐autonomous organelle, which contains its genome called plastome. Plastomes have often been used to investigate genetic differentiation and evolution of plant lineages. Plastome is smaller than mitochondrial and nuclear genomes in plant cells, and it has a large number of copies. The evolutionary rate of the chloroplast genome is higher than that of the mitochondrial genome but lower than that of the nuclear genome (Maple & Møller, 2006). The chloroplast genomes of land plants are relatively conserved in genomic structure, GC content, gene number, and gene arrangement (Xu et al., 2015). Structural variations have also been reported in diverse plant species, such as contraction and expansion in the IR regions, the loss of one IR, mixed presence of linear and circular genome, insertion or deletion, the loss of genes etc (Abdullah, Mehmood, et al., 2021; Abdullah, Shahzadi, et al., 2021; Alqahtani & Jansen, 2021; Lee et al., 2021; Li, Luo, et al., 2021; Li, Yang, et al., 2021; Sibbald & Archibald, 2020; Tonti‐Filippini et al., 2017). These characteristics of the chloroplast genome allow for its wide application in the research of evolutionary biology and molecular phylogeny, especially in the analysis of phylogenetic relationships at the intraspecific level.
In the current work, a total number of 10 newly sequenced L. obtusiloba chloroplast genomes with three published chloroplast genome sequences were compiled, which were classified into two populations. Geographically, the 13 samples allowed us (1) to evaluate the potential intraspecific variations in L. obtusiloba; (2) to assess whether the selection of the two varieties of L. obtusiloba is appropriate and sufficient for understanding the intraspecific relationship of this species, and (3) to reconstruct phylogeny between the Northern and Southern populations of L. obtusiloba. Based on the comparative chloroplast genome research, we expect to find considerable intraspecific divergence and extend our understanding of the complexity of the plastid divergence in widely distributed species.
2. MATERIALS AND METHODS
2.1. Sampling, DNA extraction, and chloroplast genome sequencing
Fresh leaves from 10 individual trees of L. obtusiloba var. obtusiloba representing local populations were collected and silica‐gel dried (Table 1). The 10 samples include two individuals collected from Northeast China (the Northern population) and eight from Southeast to Southwest China (the Southern population) (Figure 1a). Voucher specimens were deposited in the herbarium of the School of Life Sciences, Henan Agricultural University, and the Herbarium of the Xishuangbanna Tropical Botanical Garden (HITBC), Chinese Academy of Sciences. Total genomic DNA was isolated from dry leaves using the Tiangen Plant Genomic DNA Kit (Tiangen Inc., China). DNA purity was examined using Qubit 2.0 (Invitrogen Inc., USA) and NanoDrop (Thermo Scientific Inc., USA). Purified total DNA was used to make DNA libraries using the Illumina Paired‐End DNA Library Kit and sequenced with the NovaSeq 6000 platform, a service provided by NovoGene Bio‐Tech (Beijing, China). Raw reads were obtained with an average length of 150 bp, yielding at least 4 GB of clean data for each sample. The sequence data of three formerly published chloroplast genomes of L. obtusiloba were retrieved from the NCBI database and included in this analysis, with two individuals belonging to the Northern population from Liaoning, China (GenBank No. MH220737) (Zhao et al., 2018) and Korea (GenBank No. MG581448) (Jo et al., 2019), respectively, and one individual of var. heterophylla from Tibet (GenBank No. MT621623) (Li, Luo, et al., 2021; Li, Yang, et al., 2021) representing the Southern population.
TABLE 1.
Features of the ten Lindera obtusiloba new sequenced plastomes in the present study.
| Distribution | Specimen number | GenBank no. | Genome size (bp) | Large single copy (LSC; bp) | Inverted repeats (IR; bp) | Small single copy (SSC; bp) | GC content (LSC/IR/SSC) | Number of genes (CDS/tRNA/rRNA) | |
|---|---|---|---|---|---|---|---|---|---|
| Northern population | Yantai, Shandong | txy18016 | ON520653 | 154,175 | 93,751 | 20,762 | 18,900 | 39.14% (37.92%/44.29%/33.80%) | 113 (79/30/4) |
| Dalian, Liaoning | sy8679 | ON520651 | 152,770 | 93,700 | 20,080 | 18,910 | 39.12% (37.91%/44.44%/33.80%) | 113 (79/30/4) | |
| Southern population | Shangrao, Jiangxi | txy18043 | ON520654 | 152,683 | 93,633 | 20,072 | 18,906 | 39.17% (37.97%/44.44%/33.88%) | 113 (79/30/4) |
| Longnan, Gansu | sy9339 | ON520652 | 152,343 | 93,313 | 20,066 | 18,898 | 39.22% (38.06%/44.46%/33.85%) | 113 (79/30/4) | |
| Diqing, Yunnan | sy5749 | ON520645 | 152,682 | 93,630 | 20,071 | 18,910 | 39.16% (37.97%/44.45%/33.84%) | 113 (79/30/4) | |
| Linzhi, Tibet | sy6219 | ON520649 | 152,708 | 93,651 | 20,071 | 18,915 | 39.14% (37.95%/44.45%/33.80%) | 113 (79/30/4) | |
| Linzhi, Tibet | sy7768 | ON520650 | 152,708 | 93,651 | 20,071 | 18,915 | 39.14% (37.95%/44.45%/33.80%) | 113 (79/30/4) | |
| Linzhi, Tibet | sy5779 | ON520646 | 152,708 | 93,651 | 20,071 | 18,915 | 39.14% (37.95%/44.45%/33.80%) | 113 (79/30/4) | |
| Chayu, Tibet | sy5998 | ON520647 | 152,708 | 93,636 | 20,075 | 18,922 | 39.15% (37.96%/44.44%/33.82%) | 113 (79/30/4) | |
| Chayu, Tibet | sy6045 | ON520648 | 152,708 | 93,636 | 20,075 | 18,922 | 39.15% (37.96%/44.44%/33.82%) | 113 (79/30/4) | |
FIGURE 1.

Samples location of the Lindera obtusiloba (a) and their gene maps of the complete chloroplast genome (b).
2.2. Chloroplast genome assembly and annotation
The 10 complete chloroplast genomes were de novo assembled using the GetOrganelle toolkit with a typical setting (Jin et al., 2020) with the published plastomic data of Lindera from the NCBI database as a seed file. Annotation of the newly sequenced chloroplast genomes was performed with Plastid Genome Annotator (PGA) using the published L. obtusiloba (Accession no. MG581448) and L. obtusiloba var. heterophylla (Accession no. MT621623) plastome as a reference (Qu et al., 2019). To ensure the accuracy of the annotation, the same procedure was repeated with the GeSeq program (Tillich et al., 2017), which contains the HMMER profile and the tRNAscan‐SE server. The GB2sequin program was used to prepare the NCBI submission file (Lehwark & Greiner, 2019). Both programs were implemented in the CHLOROBOX web toolbox (https://chlorobox.mpimp‐golm.mpg.de/index.html). The online program of Chloroplot was used to visualize the circular genome physical map of L. obtusiloba (Zheng et al., 2020).
2.3. Whole chloroplast genome comparison
Comparative analyses across the 13 chloroplast genomes of L. obtusiloba were performed. The MISA software was used to identify simple sequence repeats (SSRs) with the following criteria: 10, 5, 4, 3, 3, and 3 repeat units are for mono‐, di‐, tri‐, tetra‐, penta‐, and hexa‐nucleotides, respectively (Beier et al., 2017). The REPuter web program was used to detect the forward, palindrome, reverse, and complement repeated elements with a minimal length of 30 bp, an identity value of more than 90%, and a Hamming distance of 3 (Kurtz et al., 2001). The intra‐genetic variations were analyzed and plotted using the mVISTA online tools with the Shuffle‐LAGAN mode (Frazer et al., 2004). To evaluate the significance of the analyses, three datasets representing the total population, the Northern population, and the Southern population were selected and analyzed. Nucleotide diversity (Pi) for each dataset was detected using the DnaSP software (Rozas et al., 2017). Furthermore, the sliding window analysis was conducted with a reference nucleotide diversity of 0.008 with a step size of 200 bp and a window length of 400 bp.
2.4. Phylogenomic analyses
The phylogenetic relationship of individuals within L. obtusiloba and to some related species from the “core” Laureae was interpreted using two separate datasets, the whole chloroplast genome and the protein‐coding genes (PCGs). A total of 65 chloroplast genomes representing 54 species or subspecies were included in this analysis, which covers the seven genera of the “core” Laureae. The species Cinnamomum camphora (L.) Presl belonging to the genus Cinnamomum Schaeff was used as the outgroup (Appendix S1). A multiple sequence alignment was generated with MAFFT under a default setting (Katoh & Standley, 2013) and then manually adjusted in BioEdit (Hall et al., 2011). Phylogenetic trees were constructed using both Maximum likelihood (ML) and Bayesian inference (BI) methods. The best‐fit nucleotide substitution model is GTR + F + R9 for the whole plastomic sequences and GTR + F + R3 for the coding regions determined with ModelFinder using the AIC values as criteria (Kalyaanamoorthy et al., 2017). In the ML analysis, IQ‐TREE (Nguyen et al., 2014) was implemented, and a bootstrap value of 50,000 was assigned with the SH‐aLRT branch test. In the BI analysis, MrBayes (Ronquist et al., 2012) was employed with two independent Markov Chain Monte Carlo chains for 2,000,000 generations. The first 25% of trees were discarded, and the split frequency is below 0.01.
2.5. Selective pressure analysis
Forty‐five protein‐coding genes were used to search for signs of positive selection in the L. obtusiloba chloroplast genomes with EasyCodeML (Gao et al., 2019). The unrooted phylogenetic tree file of L. obtusiloba species was generated using the ML approach. The ratio (ω) of the nonsynonymous substitution rate (dN) to the synonymous substitutions rate (dS) for each protein‐coding gene was determined using both the site model and the branch model. Signatures of adaptation were analyzed using the site model with four site‐specific models (M0 vs. M3, M1a vs. M2a, M7 vs. M8, and M8a vs. M8). The branch model was adopted under the one‐ratio model (M0) and the two‐ratio model with the Northern population of L. obtusiloba as the foreground branch. In addition, a likelihood ratio test (LRT) was performed to assess the significance of these analyses (Yang, 1998).
3. RESULTS
3.1. General characteristics of L. obtusiloba chloroplast genomes
The 13 L. obtusiloba plastomes share a conserved quadripartite circular structure with an overall length ranging from 152,682 bp to 154,175 bp (Figure 1b). The plastome size of the Northern population is comparably larger than that of the Southern population (Table 1). These chloroplast genomes all contain a large single‐copy region (LSC; length: 93,313 bp to 93,751 bp) and a small single‐copy region (SSC; length: 18,898 bp to 18,922 bp), which are separated by two inverted repeat regions (IRa and IRb; length: 20,066 bp to 20,762 bp) (Table 1). All individuals from the North possess a larger LSC region when compared to the Southern population. Species from Liaoning (a city in North China) share a similar IR size with individuals from the Southern population. The individuals from Gansu, which belongs to the Southern population, have the smallest SSC region. All individuals from Tibet share similar genome sizes, LSCs, IRs, and SSCs. In addition, the GC contents of all sampled plastomes vary slightly between 39.12% to 39.22%. Moreover, the GC content of IRs (44.29–44.45%) was higher than that of LSC (38.06–37.96%) and SSC (33.80–33.88%) regions. All the analyzed chloroplast genomes harbor 113 unique genes, including 79 PCGs, 30 tRNAs, and 4 rRNAs. Five PCGs and 6 tRNA genes are duplicated in the IR regions. The rps12 gene was identified as a trans‐spliced gene with two duplicates in the IR regions and one in the LSC region.
3.2. Comparative phylogenomic analyses of L. obtusiloba plastomes
In this study, comparative analyses of the 13 plastomes were performed. A total of 371 longer dispersed repeats were detected and classified into 4 categories including forward, reverse, complement, and palindromic direct match repeats (Appendix S2). All individuals have a similar number of longer dispersed repeats, ranging from 20 to 27. The number of forward and palindromic repeats is 11–13 and 12–15, respectively. Among all the analyzed plastomes, the Northern L. obtusiloba individuals do not have reverse repeats except for the sample from Shandong, which has one reverse repeat but no complement repeats (Figure 2a). A total of 807 SSRs were identified among all species, which fall into 5 classes: mono‐, di‐, tri‐, tetra‐, and hexa‐nucleotide repeats (Appendix S2). All of the Northern individuals contain a larger number of mono‐nucleotide repeats than that of the Southern population (Figure 2b). The number of mono‐, di‐, tri‐, and tetra‐nucleotides is 52, 8, 2, and 6, respectively, for the Northern population, while it is 49, 9, 1, and 6 for spices from Shandong (Appendix S2). Among the Southern population, individuals from Gansu contain all five classes of repeats, while the number of mono‐nucleotide repeats (18) is relatively low. It is notable that the individuals from Yunnan only have mono‐ and di‐nucleotide repeats. Among the SSRs, the mono‐nucleotide repeats are the most frequent type with a typical A/T repeat unit, followed by the di‐nucleotide repeats, which are composed of TA/AT and/or GA/TC. The most commonly observed tetra‐nucleotide repeat units are AAAT/TTTA, AACT, AATG/CATT, ATAC, CATA, TTAT, and TTTC. Tri‐ (AAT and TAT) and hexa‐nucleotide (TAGAAT and TTCTAT) repeats are relatively rare (Appendix S2).
FIGURE 2.
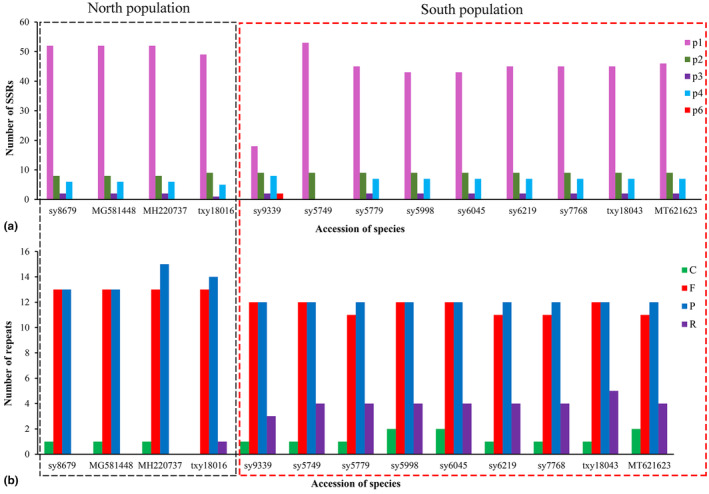
Number of repeats in the thirteen Lindera obtusiloba chloroplast genomes. (a) Total number of SSRs detected in each species. (b) Total number of the forward (F), palindrome (P), reverse (R), and complement (C) repeated elements detected in each species.
Nucleotide variations among intra‐ and inter‐populations of L. obtusiloba were also investigated in this work. The comparison suggests a high level of divergence among populations but a low level of divergence within a given population of L. obtusiloba (Table 2). A total of 329 singleton variable sites, 371 parsimony informative sites, and 340 InDels were identified across the whole chloroplast genomes with a nucleotide diversity value (π) of 0.00136. The coding regions are more conserved than the noncoding regions, with a π of 0.00118, which includes 141 singleton variable sites, 167 parsimony informative sites, and 71 InDels. Nucleotide diversity level was highly similar between the Northern (π = 0.00075 and 0.00066) and the Southern (π = 0.00076 and 0.00065) populations across the whole chloroplast genomes and the PCGs. However, the proportion of parsimony informative sites and InDels in the Southern population is higher than that of the Northern population (Table 2). Hypervariable regions of all L. obtusiloba were identified by the slide window test (Figure 3). Four regions possessing a nucleotide diversity larger than 0.008 were found, which are trnH ‐GUG‐psbA, rpoC2‐rpoC1, ycf1‐ndhF, and ccsA‐ndhD. In the Northern individuals, rpoC2‐rpoC1 and ccsA‐ndhD show higher nucleotide diversity, while variable regions were not observed in the Southern individuals. With the mVISTA program, the sequence divergence of L. obtusiloba among all examined individuals was detected and analyzed with the Lavandula latifolia Hook. f. sequence as reference (Figure 4), the whole chloroplast genomes are conserved in structure and gene order. The non‐coding regions are more likely to accumulate variations than the coding regions, and IR regions are more conserved than the LSC and SSC regions.
TABLE 2.
Comparison of the variable sites of the 13 Lindera obtusiloba plastomes.
| Sequences | Variables | North population | South population | Total |
|---|---|---|---|---|
| Whole genome sequence | Singleton variable sites | 229 | 238 | 329 |
| Parsimony informative sites | 1 | 142 | 371 | |
| Nucleotide diversity | 0.00075 | 0.00076 | 0.00136 | |
| InDels events | 99 | 215 | 340 | |
| Protein coding sequence | Singleton variable sites | 102 | 108 | 141 |
| Parsimony informative sites | 1 | 59 | 167 | |
| Nucleotide diversity | 0.00066 | 0.00065 | 0.00118 | |
| InDels events | 24 | 43 | 71 |
FIGURE 3.

Sliding window analysis of the entire chloroplast genome of thirteen Lindera obtusiloba individuals (window length: 600 bp; step size: 200 bp). x‐axis: position of the midpoint of a window; y‐axis: nucleotide diversity of each window.
FIGURE 4.

Visualization of genome alignment of the thirteen complete chloroplast genomes from Lindera obtusiloba. The cp genome of Lavandula latifolia is used as the reference. x‐axis indicates the sequence coordinates in the whole cp genome. y‐axis represents the similarity of the aligned regions, indicating percent identity to the reference genome (50–100%).
3.3. Phylogenetic analysis of L. obtusiloba within the “core Laureae”
To gain more insights into the relationship among various L. obtusiloba individuals and other closely related species, a total of 65 chloroplast genomes including 13 L. obtusiloba individuals and other 51 “core Laureae” species were used for further phylogenetic analysis, which was conducted with both ML and BI methods using both the complete chloroplast genomes and the shared protein‐coding genes.
Independent of the data and the inference method used, the obtained trees share a similar topology except for the Lindera IV‐VI clades, which contain the Lindera sect. Aperula, sect. Polyadenia, sect. Lindera, respectively (Figure 5). In the plastome‐based phylogeny, the Lindera IV clade represents a sister lineage of a larger group containing the genus Parasassafras, the North America Lindera species (Lindera III), genus Neolitsea‐Actinodaphne (Neo and Act complex) and the trinerved Lindera complex (Lindera II) (ML analysis; Figure 5). However, according to the BI analysis, Lindera IV is grouped with Parasassafras first and then shares a sister relationship with other clades (Appendix S3). When only protein‐coding genes are used, the BI tree is similar in topology to the plastome‐based ML tree (Appendix S3). However, the ML analysis with the coding genes predicts a sister relationship between the clade containing Parasassafras and other clades including the Lindera IV clade (Appendix S3). A sister relationship between clade Lindera V and VI was strongly supported by all phylogenetic trees, while the phylogenetic location of these two clades was inconsistent according to analyses with the whole genome sequences and protein‐coding sequences. Based on the whole chloroplast genome analysis, they formed a larger clade with Litsea I, Lindera I to IV, Parasassafras, and the Neo and Act complex (Figure 5; Appendix S3), but are closer to the clade containing genus Litsea (Litsea II and III), L. praecox (Siebold and Zucc.) Blume from sect. Lindera and L. rubronervia Gamble from sect. Sphaerocarpae (Lindera VII), genus Laurus L. when using protein‐coding genes (Appendix S3).
FIGURE 5.

Phylogenetic tree reconstruction of ‘core’ Laureae using the maximum likelihood (ML) method based on complete chloroplast genome sequences. Only the ML tree is shown, because its topology is identical to that of the obtained BI tree.
The results all support a close relationship between L. obtusiloba (Lindera I) and the genus Litsea (Litsea I), and that the individuals have diverged into the Northern and the Southern lineages (Figure 5). However, analyses with both datasets failed to produce a species boundary between L. obtusiloba var. obtusiloba and L. obtusiloba var. heterophylla. Although the Northern population and the Southern population are separated in the phylogenetic trees, the L. obtusiloba individuals in each region are still clustered in a mixed way on the obtained tees.
3.4. Selection analyses
Most protein‐coding genes are under purifying selection indicated by their low dN/dS ratios in the analyses with both the branch model and the site model. Five genes (atpB, ndhC, rpl16, petD, rpoC2) were found to have evolved under positive selection in the L. obtusiloba clade (Figure 5) according to Site model analysis. It was found that four sites were potentially under positive selection in petD and rpoC2 and that only a single site was detected for atpB, ndhC, and rpl16 (p < .05) (Appendix S4). Seven amino acid differences were observed between the Northern and the Southern populations of L. obtusiloba for rpoC2 (Figure 6). The Branch model analysis supports strong selective pressure on clpP, petB, rpl16, and rpoC1, which put the Northern individuals into the foreground branch and the Southern individuals into the background branch (Appendix S4). However, the evolutionary significance of these sites cannot be accurately evaluated at this point due to the lack of 3D structures and functional analysis of these proteins.
FIGURE 6.

Phylogenetic tree reconstruction and positive selection genes of Lindera obtusiloba using the maximum likelihood (ML) method based on 79 coding sequences. Genes were colored with blue background were identified by site model, Genes were colored with red background were identified by branch model with the Northern population of L. obtusiloba as the foreground branch. Amino acid differences were observed between the Northern and the Southern populations of L. obtusiloba for rpoC2 gene. *means p < 0.05 according selection analyses. **means p <0.01 according selection analyses.
4. DISCUSSION
In this study, intraspecific variations of L. obtusiloba were examined by analyzing 13 plastomes, covering the Northern and Southern geographic types. Despite their similarities in overall structure, gene order, and GC contents, the plastomes do vary in size, with L. obtusiloba Shandong being the largest. The variations in size could be attributed to the expansion of the IR regions in this sample (Abdullah, Mehmood, et al., 2021; Abdullah, Shahzadi, et al., 2021; Zhang et al., 2020). The chloroplast genome of L. obtusiloba from Shandong has a 678‐nucleotide insertion in its IR regions, which explains why it is bigger than other individuals (Table 2). A longer LSC with multiple InDels in the Northern population might also account for the size variation. These results reflect that both expansions in the IRs and InDel events can lead to variations in the plastomes at the intraspecific level (Jiang et al., 2017; Lei et al., 2016; Muraguri et al., 2020). Although the Northern samples contain larger chloroplast genomes, they have fewer parsimony informative sites and InDels when compared to the Southern individuals (Table 2), indicating a shorter evolutionary history and relatively homogeneous topography in North of East Asia (Tian et al., 2020). This is also supported by the fact that the Northern individuals possess less diversity in the SSR types and the number of repeats than the Southern population (Tian et al., 2020), which contradicts the results of a previous study by Ye et al. (2017). This difference may be related to the number of individuals and DNA target loci selected. Due to the presence of intraspecific variations, more representative populations need to be included when investigating genome diversity and intraspecific boundaries of species with a broad range of distribution.
The mVISTA results suggest that the coding regions are highly conserved and that variations are more likely to appear in the LSC and SSC regions than in the IR regions. This is consistent with previous findings with other Lauraceae species and flowering plants (Li, Luo, et al., 2021; Li, Yang, et al., 2021; Liu, Chen, et al., 2021; Liu, Ma, et al., 2021; Song et al., 2020; Yang et al., 2022). A total of four intergenic variable regions were identified across all the analyzed chloroplast genomes. Among these, two highly variable regions were found in the Northern population, but no high variable regions were detected in individuals from the Southern populations (Figure 3). It is speculated that the difference in cpDNA divergence patterns between the Northern and the Southern populations might be shaped by their specific evolutionary history (Tian et al., 2020). For the Northern populations, most species have a recent population history, which was formed via postglacial migration, while Southern populations may have served as glacial refugia during Pleistocene climate fluctuations (Milne & Abbott, 2002; Qiu et al., 2011; Ye et al., 2017). Additionally, the highly variable regions discovered in this study contain more variable sites when compared with one previous study (Ye et al., 2017). Based on their study, the psbA–trnH, trnL–trnF, trnS–trnG, and rpl16 chloroplast DNA markers were designed. The variable regions detected in this study can also be used as potential DNA markers for studying the evolutionary history of L. obtusiloba and resolving the taxonomic status of L. obtusiloba with global sampling.
Phylogenetic trees were reconstructed using the chloroplast genomes of L. obtusiloba from different geographic regions and 51 plastomes of the “core Laureae.” Our results support the monophyletic origin of the “core Laureae” and the monophyletic status of the clade Litsea I, Lindera II, Lindera IV, Lindera V, and Lindera VI, which are consistent with some previous work (Jo et al., 2019; Liu, Chen, et al., 2021; Liu, Ma, et al., 2021; Song et al., 2020; Tian et al., 2019; Xiao et al., 2020). Phylogenetic results of the clade Litsea III, Lindera V, and Lindera VI in our studies are in agreement with a previous phylogenetic work by Zhao et al. (2018), who defined the relationships among the species located in sub‐clade I, and sub‐clade II containing Lindera sect. Aperula. However, the species, L. benzoin (L.) Blume distributed in North America showed a close relationship with the Lindera sect. Daphnidium, genus Neolitsea, and Actinodaphne (Figure 5), which is inconsistent with Zhao et al. (2018). This inconsistency can be explained by the difference in the samples used in phylogenetic reconstruction. In this study, the species of L. obtusiloba were placed in one of the Litsea clades (Litsea I; Figure 5), which agrees with the previous studies (Fijridiyanto & Murakami, 2009; Li et al., 2004; Liu, Chen, et al., 2021; Liu, Ma, et al., 2021; Song et al., 2020). L. obtusiloba usually has trifid, pentafidentire, or sometimes entire leaf margins, and pentanerved or trinerved veins, which is significantly different from other Lindera plants (Li, Conran, et al., 2008; Li, Li, & Li, 2008; Tian et al., 2020; Ye et al., 2017). The differences in the shape, arrangement, and pubescence of leaves may have a prominent influence on the rates of photosynthesis and transpiration, which might have allowed some variety to become better adapted to its particular environment (Chitwood & Sinha, 2016; Ding et al., 2020). In our study, five genes were identified with positively selected sites. These genes include the ATP subunit gene (atpB), NAD(P)H dehydrogenase complex gene (ndhC), small subunits of ribosome gene (rpl16), cytochrome b/f complex subunit proteins (petD), and DNA‐dependent RNA polymerase gene (rpoC2). Additionally, rpoC2 is one of the largest chloroplast genes, encoding the RNA polymerase β, which is essential for the transcription of several photosynthesis‐related genes (Cummings et al., 1994). This gene possesses four positively selected sites, and seven nonsynonymous substitutions were detected at seven other sites (Figure 6). L. obtusiloba is the northernmost species of the family Lauraceae, which is highly sensitive to aridity, and it has evolved adaptively to its surrounding environments, presumably through the regulation of photosynthesis‐related genes and the modification of the photosynthetic system. Based on the phylogeny, L. obtusiloba shares a close relationship with the genus Litsea. However, there is no obvious morphological evidence supporting their close relationship (Li, Conran, et al., 2008; Li, Li, & Li, 2008). As a result, further study at both the molecular and morphological levels with more intensive sampling will be necessary.
The species boundary of L. obtusiloba between var. obtusiloba and var. heterophylla is unresolved in the current work. As a result, we inferred that gene flow might still occur between these two varieties, which is supported by the presence of multiple refuges in the Hengduan Mountains (Ye et al., 2017). In the current work, four photosynthesis‐related genes that are positively selected were identified in the Southern population, which suggests that environmental heterogeneity might have led to more intraspecific variations in this particular population (Tian et al., 2020). Together, these make it extremely challenging to distinguish the two varieties in the Southwest region of East Asia. Consequently, further examination potentially involving nuclear genomic data will be critical to fully elucidate the possibility of having hybridization or gene flow occur in the individuals from the admixture regions.
5. CONCLUSIONS
In this study, we investigated the intraspecific diversity of L. obtusiloba by comparative plastomic and phylogenetic analyses. The structural characteristics, gene content, nucleotide variations, InDel events, and sequence repeats of the 13 L. obtusiloba chloroplast genomes are highly conserved. Here, two groups congruent with the geographical distribution were found, and the aridity belt is responsible for this intraspecific divergence. Differences in nucleotide variable sites, simple sequence repeats, and variable hotspot regions support the discontinuous distribution of L. obtusiloba in Northern and Southern East Asia. L. obtusiloba in different regions has experienced different evolutionary pressures posed by various geographical, environmental, historical, and climatic factors, which have evolved a few photosynthesis‐related genes adaptively under positive selection. The phylogenetic confirmed the position of L. obtusiloba within the “core Laureae” and suggests that the relationship between L. obtusiloba, and genus Litsea should be provided more evidence. However, considering its wide distribution, a larger number of chloroplast and nuclear genomes should be included in further analysis to fully understand the evolutionary history of L. obtusiloba.
AUTHOR CONTRIBUTIONS
Xiangyu Tian: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); resources (lead); software (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Jia Guo: Data curation (equal); funding acquisition (equal); investigation (equal); validation (equal); writing – review and editing (equal). Yu Song: Conceptualization (equal); funding acquisition (equal); project administration (lead); resources (lead); supervision (equal); writing – review and editing (equal). Qunfei Yu: Data curation (equal); formal analysis (equal); resources (equal). Cao Liu: Data curation (equal); formal analysis (equal); methodology (equal); resources (equal); visualization (equal). Zhixi Fu: Funding acquisition (equal); project administration (equal); visualization (equal); writing – review and editing (equal). Yuhua Shi: Investigation (equal); resources (equal); validation (equal); writing – review and editing (equal). Yizhen Shao: Methodology (equal); resources (equal); software (equal); validation (equal). Zhiliang Yuan: Conceptualization (equal); funding acquisition (equal); project administration (equal); resources (equal); supervision (equal); writing – review and editing (equal).
FUNDING INFORMATION
This work was partially supported by grants from the Applied Basic Research Project of Yunnan (2019FB057), the National Natural Science Foundation of China (No. 32000158), and the Technologies Research and Development Program of Henan (No. 222102110009).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest for the publication of the manuscript.
Supporting information
Appendix S1.
Appendix S2.
Appendix S3.
Appendix S4.
ACKNOWLEDGEMENTS
We are grateful to Hong‐fang Wang from Beijing Normal University, Jun‐wei Ye from Southwest Forestry University, and Yong‐sheng Chen from Liaoning Normal University for their constructive comments regarding the manuscript. We are also indebted to anonymous reviewers and the editor for their constructive comments and suggestions, which have helped to improve this work.
Tian, X. , Guo, J. , Song, Y. , Yu, Q. , Liu, C. , Fu, Z. , Shi, Y. , Shao, Y. , & Yuan, Z. (2024). Intraspecific differentiation of Lindera obtusiloba as revealed by comparative plastomic and evolutionary analyses. Ecology and Evolution, 14, e11119. 10.1002/ece3.11119
Contributor Information
Yu Song, Email: songyu@gxnu.edu.cn.
Zhiliang Yuan, Email: zhiliangyuan@henau.edu.cn.
DATA AVAILABILITY STATEMENT
The Lindera obtusiloba plastomes generated in this study are available in the NCBI GenBank repository with accession numbers ON520645 to ON520654. The Supplementary data to this article can be found online at https://10.1002/ece3.11119.
REFERENCES
- Abdullah, F. M. , Shahzadi, I. , Ali, Z. , Islam, M. , Naeem, M. , Mirza, B. , Lockhart, P. J. , Ahmed, I. , & Waheed, M. T. (2021). Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. Journal of Systematics and Evolution, 59(2), 388–402. [Google Scholar]
- Abdullah, M. , Mehmood, F. , Heidari, P. , Rahim, A. , Ahmed, I. , & Poczai, P. (2021). Pseudogenization of the chloroplast threonine (trnT‐GGU) gene in the sunflower family (Asteraceae). Scientific Reports, 11(1), 21122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix, K. , Gérard, P. R. , Schwarzacher, T. , & Heslop‐Harrison, J. S. P. (2017). Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Annals of Botany, 120(2), 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani, A. A. , & Jansen, R. K. (2021). The evolutionary fate of rpl32 and rps16 losses in the Euphorbia schimperi (Euphorbiaceae) plastome. Scientific Reports, 11(1), 7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschehoug, E. T. , Brooker, R. , Atwater, D. Z. , Maron, J. L. , & Callaway, R. M. (2016). The mechanisms and consequences of interspecific competition among plants. Annual Review of Ecology, Evolution, and Systematics, 47(1), 263–281. [Google Scholar]
- Bai, W. N. , Wang, W. T. , & Zhang, D. Y. (2016). Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytologist, 209(4), 1757–1772. [DOI] [PubMed] [Google Scholar]
- Beier, S. , Thiel, T. , Münch, T. , Scholz, U. , & Mascher, M. (2017). MISA‐web: A web server for microsatellite prediction. Bioinformatics, 33(16), 2583–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist, D. , Pauliny, A. , Larsson, M. , & Flodin, L. Å. (2010). Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evolutionary Biology, 10(1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger, R. , Schneider, K. A. , & Willensdorfer, M. (2006). The conditions for speciation through intraspecific competition. Evolution, 60(11), 2185–2206. [PubMed] [Google Scholar]
- Cao, Y. , Xuan, B. , Peng, B. , Li, C. , Chai, X. , & Tu, P. (2016). The genus Lindera: A source of structurally diverse molecules having pharmacological significance. Phytochemistry Reviews, 15(5), 869–906. [Google Scholar]
- Chanderbali, A. , Van der Werff, H. , & Renner, S. (2001). Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Annals of the Missouri Botanical Garden, 88(1), 104–134. [Google Scholar]
- Chen, T. Y. , & Lou, A. R. (2019). Phylogeography and paleodistribution models of a widespread birch (Betula platyphylla Suk.) across East Asia: Multiple refugia, multidirectional expansion, and heterogeneous genetic pattern. Ecology and Evolution, 9, 7792–7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D. H. , & Sinha, N. R. (2016). Evolutionary and environmental forces sculpting leaf development. Current Biology, 26(7), R297–R306. [DOI] [PubMed] [Google Scholar]
- Cummings, M. P. , King, L. M. , & Kellogg, E. A. (1994). Slipped‐strand mispairing in a plastid gene: rpoC2 in grasses (Poaceae). Molecular Biology and Evolution, 11(1), 1–8. [DOI] [PubMed] [Google Scholar]
- Des Roches, S. , Post, D. M. , Turley, N. E. , Bailey, J. K. , Hendry, A. P. , Kinnison, M. T. , Schweitzer, J. A. , & Palkovacs, E. P. (2018). The ecological importance of intraspecific variation. Nature Ecology & Evolution, 2(1), 57–64. [DOI] [PubMed] [Google Scholar]
- Díaz, S. , Pascual, U. , Stenseke, M. , Martín‐López, B. , Watson, R. T. , Molnár, Z. , Hill, R. , Chan, K. M. A. , Baste, I. A. , Brauman, K. A. , Polasky, S. , Church, A. , Lonsdale, M. , Larigauderie, A. , Leadley, P. W. , van Oudenhoven, A. P. E. , van der Plaat, F. , Schröter, M. , Lavorel, S. , … Shirayama, Y. (2018). Assessing nature's contributions to people. Science, 359(6373), 270–272. [DOI] [PubMed] [Google Scholar]
- Ding, J. Y. , Johnson, E. A. , & Martin, Y. E. (2020). Optimization of leaf morphology in relation to leaf water status: A theory. Ecology and Evolution, 10(3), 1510–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijridiyanto, I. , & Murakami, N. (2009). Phylogeny of Litsea and related genera (Laureae‐Lauraceae) based on analysis of rpb2 gene sequences. Journal of Plant Research, 122(3), 283–298. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A. , Pachter, L. , Poliakov, A. , Rubin, E. M. , & Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Research, 32, W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freise, C. , Erben, U. , Neuman, U. , Kim, K. , Zeitz, M. , Somasundaram, R. , & Ruehl, M. (2010). An active extract of Lindera obtusiloba inhibits adipogenesis via sustained Wnt signaling and exerts anti‐inflammatory effects in the 3T3‐L1 preadipocytes. The Journal of Nutritional Biochemistry, 21(12), 1170–1177. [DOI] [PubMed] [Google Scholar]
- Gao, F. , Chen, C. , Arab, D. A. , du, Z. , He, Y. , & Ho, S. Y. W. (2019). EasyCodeML: A visual tool for analysis of selection using CodeML. Ecology and Evolution, 9(7), 3891–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, L. N. , Barley, A. J. , Poe, S. , Thomson, R. C. , Nieto‐Montes de Oca, A. , & Wang, I. J. (2019). Phylogeography of a widespread lizard complex reflects patterns of both geographic and ecological isolation. Molecular Ecology, 28(3), 644–657. [DOI] [PubMed] [Google Scholar]
- Guo, Z. , Sun, B. , Zhang, Z. , Peng, S. , Xiao, G. , Ge, J. , Hao, Q. , Qiao, Y. , Liang, M. , Liu, J. , Yin, Q. , & Wei, J. J. (2008). A major reorganization of Asian climate by the early Miocene. Climate of the Past, 4(3), 153–174. [Google Scholar]
- Halbritter, A. H. , Fior, S. , Keller, I. , Billeter, R. , Edwards, P. J. , Holderegger, R. , Karrenberg, S. , Pluess, A. R. , Widmer, A. , & Alexander, J. M. (2018). Trait differentiation and adaptation of plants along elevation gradients. Journal of Evolutionary Biology, 31(6), 784–800. [DOI] [PubMed] [Google Scholar]
- Hall, T. , Biosciences, I. , & Carlsbad, C. (2011). BioEdit: An important software for molecular biology. GERF Bulletin of Biosciences, 2(1), 60–61. [Google Scholar]
- Hong, C. O. , Rhee, C. H. , Won, N. H. , Choi, H. D. , & Lee, K. W. (2013). Protective effect of 70% ethanolic extract of Lindera obtusiloba Blume on tert‐butyl hydroperoxide‐induced oxidative hepatotoxicity in rats. Food and Chemical Toxicology, 53, 214–220. [DOI] [PubMed] [Google Scholar]
- Huang, C. L. , Chen, J. H. , Chang, C. T. , Chung, J. D. , Liao, P. C. , Wang, J. C. , & Hwang, S. Y. (2016). Disentangling the effects of isolation‐by‐distance and isolation‐by‐environment on genetic differentiation among rhododendron lineages in the subgenus Tsutsusi. Tree Genetics & Genomes, 12(3), 53. [Google Scholar]
- Jiang, D. , Zhao, Z. , Zhang, T. , Zhong, W. , Liu, C. , Yuan, Q. , & Huang, L. (2017). The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes, 8(9), 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. J. , Yu, W. B. , Yang, J. B. , Song, Y. , de Pamphilis, C. W. , Yi, T. S. , & Li, D. Z. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 21(1), 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, Y. K. , Cheon, S. H. , Fan, Q. , & Kim, K. J. (2019). Characterization of 20 complete plastomes from the tribe Laureae (Lauraceae) and distribution of small inversions. PLoS One, 14(11), e0224622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy, S. , Minh, B. Q. , Wong, T. K. F. , von Haeseler, A. , & Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14(6), 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, S. , Choudhuri, J. V. , Ohlebusch, E. , Schleiermacher, C. , Stoye, J. , & Giegerich, R. (2001). REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research, 29(22), 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. , Choi, I.‐S. , Cardoso, D. , de Lima, H. C. , de Queiroz, L. P. , Wojciechowski, M. F. , Jansen, R. K. , & Ruhlman, T. A. (2021). The chicken or the egg? Plastome evolution and an independent loss of the inverted repeat in papilionoid legumes. The Plant Journal, 107(3), 861–875. [DOI] [PubMed] [Google Scholar]
- Lehwark, P. , & Greiner, S. (2019). GB2sequin ‐ a file converter preparing custom GenBank files for database submission. Genomics, 111(4), 759–761. [DOI] [PubMed] [Google Scholar]
- Lei, W. , Ni, D. , Wang, Y. , Shao, J. , Wang, X. , Yang, D. , Wang, J. , Chen, H. , & Liu, C. (2016). Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Scientific Reports, 6(1), 21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, D. M. , Hendry, A. P. , Vázquez‐Domínguez, E. , & Friesen, V. L. (2019). Estimated six per cent loss of genetic variation in wild populations since the industrial revolution. Evolutionary Applications, 12(8), 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. T. , Luo, Y. , Gan, L. , Ma, P. F. , Gao, L. M. , Yang, J. B. , Cai, J. , Gitzendanner, M. A. , Fritsch, P. W. , Zhang, T. , Jin, J. J. , Zeng, C. X. , Wang, H. , Yu, W. B. , Zhang, R. , van der Bank, M. , Olmstead, R. G. , Hollingsworth, P. M. , Chase, M. W. , … Li, D. Z. (2021). Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biology, 19(1), 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. W. (1985). Parallel evolution in Listea and Lindera of Lauraceae. Acta Botanica Yunnanica, 7(2), 129–135. [Google Scholar]
- Li, J. , Christophel, D. , Conran, J. , & Li, H. W. (2004). Phylogenetic relationships within the ‘core’ Laureae (Litsea complex, Lauraceae) inferred from sequences of the chloroplast gene matK and nuclear ribosomal DNA ITS regions. Plant Systematics and Evolution, 246(1–2), 19–34. [Google Scholar]
- Li, J. , Conran, J. G. , Christophel, D. C. , Li, Z. M. , Li, L. , & Li, H. W. (2008). Phylogenetic relationships of the Litsea Complex and Core Laureae (Lauraceae) using ITS and ETS sequences and morphology. Annals of the Missouri Botanical Garden, 95, 580–599. [Google Scholar]
- Li, L. , Li, J. , Conran, J. G. , Li, X. W. , & Li, H. W. (2007). Phylogeny of Neolitsea (Lauraceae) inferred from Bayesian analysis of nrDNA ITS and ETS sequences. Plant Systematics and Evolution, 269(3–4), 203–221. [Google Scholar]
- Li, S. G. , Li, X. W. , & Li, J. (2008). Lauraceae. In Wu Z. Y., Raven P. H., & Hong D. Y. (Eds.), Flora of China (Vol. 7, pp. 102–254). Science Press, Beijing & Missouri Botanical Garden Press. [Google Scholar]
- Li, X. , Yang, J. B. , Wang, H. , Song, Y. , Corlett, R. T. , Yao, X. , Li, D. Z. , & Yu, W. B. (2021). Plastid NDH Pseudogenization and gene loss in a recently derived lineage from the largest Hemiparasitic plant genus Pedicularis (Orobanchaceae). Plant and Cell Physiology, 62(6), 971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Chen, H. , Tang, L. , Khine, P. K. , Han, L. H. , Song, Y. , & Tan, Y. H. (2021). Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae). Plant Diversity, 44(4), 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. F. , Ma, H. , Ci, X. Q. , Li, L. , Song, Y. , Liu, B. , Li, H. W. , Wang, S. L. , Qu, X. J. , Hu, J. L. , Zhang, X. Y. , Conran, J. G. , Twyford, A. D. , Yang, J. B. , Hollingsworth, P. M. , & Li, J. (2021). Can plastid genome sequencing be used for species identification in Lauraceae? Botanical Journal of the Linnean Society, 197(1), 1–14. [Google Scholar]
- Maple, J. , & Møller, S. G. (2006). Plastid division: Evolution, mechanism and complexity. Annals of Botany, 99(4), 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Serra, A. , & Benson, R. B. J. (2020). Developmental constraints do not influence long‐term phenotypic evolution of marsupial forelimbs as revealed by interspecific disparity and integration patterns. The American Naturalist, 195(3), 547–560. [DOI] [PubMed] [Google Scholar]
- Milne, R. I. , & Abbott, R. J. (2002). The origin and evolution of tertiary relict floras. Advances in Botanical Research, 38, 281–314. [Google Scholar]
- Mimura, M. , Yahara, T. , Faith, D. P. , Vázquez‐Domínguez, E. , Colautti, R. I. , Araki, H. , Javadi, F. , Núñez‐Farfán, J. , Mori, A. S. , Zhou, S. , Hollingsworth, P. M. , Neaves, L. E. , Fukano, Y. , Smith, G. F. , Sato, Y. I. , Tachida, H. , & Hendry, A. P. (2017). Understanding and monitoring the consequences of human impacts on intraspecific variation. Evolutionary Applications, 10(2), 121–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraguri, S. , Xu, W. , Chapman, M. , Muchugi, A. , Oluwaniyi, A. , Oyebanji, O. , & Liu, A. (2020). Intraspecific variation within Castor bean (Ricinus communis L.) based on chloroplast genomes. Industrial Crops and Products, 155, 112779. [Google Scholar]
- Nguyen, L. T. , Schmidt, H. A. , von Haeseler, A. , & Minh, B. Q. (2014). IQ‐TREE: A fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. X. , Fu, C. X. , & Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of quaternary climate and environmental change in the world's most diverse temperate flora. Molecular Phylogenetics and Evolution, 59(1), 225–244. [DOI] [PubMed] [Google Scholar]
- Qu, X. J. , Moore, M. J. , Li, D. Z. , & Yi, T. S. (2019). PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15(1), 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer, J. G. (2000). Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Systematic Botany, 25, 60–71. [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D. L. , Darling, A. , Höhna, S. , Larget, B. , Liu, L. , Suchard, M. A. , & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J. , Ferrer‐Mata, A. , Sánchez‐DelBarrio, J. C. , Guirao‐Rico, S. , Librado, P. , Ramos‐Onsins, S. E. , & Sánchez‐Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12), 3299–3302. [DOI] [PubMed] [Google Scholar]
- Ruckelshaus, M. H. , Jackson, S. T. , Mooney, H. A. , Jacobs, K. L. , Kassam, K. A. S. , Arroyo, M. T. K. , Báldi, A. , Bartuska, A. M. , Boyd, J. , Joppa, L. N. , Kovács‐Hostyánszki, A. , Parsons, J. P. , Scholes, R. J. , Shogren, J. F. , & Ouyang, Z. (2020). The IPBES global assessment: Pathways to action. Trends in Ecology & Evolution, 35(5), 407–414. [DOI] [PubMed] [Google Scholar]
- Ruhlman, T. A. , & Jansen, R. K. (2014). The plastid genomes of flowering plants. In Chloroplast biotechnology (pp. 3–38). Springer. [DOI] [PubMed] [Google Scholar]
- Sibbald, S. J. , & Archibald, J. M. (2020). Genomic insights into plastid evolution. Genome Biology and Evolution, 12(7), 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Yu, W.‐B. , Tan, Y.‐H. , Jin, J.‐J. , Wang, B. , Yang, J.‐B. , Liu, B. , & Corlett, R. T. (2020). Plastid Phylogenomics improve phylogenetic resolution in the Lauraceae. Journal of Systematics and Evolution, 58(4), 423–439. [Google Scholar]
- Taudt, A. , Colomé‐Tatché, M. , & Johannes, F. (2016). Genetic sources of population epigenomic variation. Nature Reviews Genetics, 17(6), 319–332. [DOI] [PubMed] [Google Scholar]
- Tian, X. Y. , Ye, J. W. , & Song, Y. (2019). Plastome sequences help to improve the systematic position of Trinerved Lindera species in the family Lauraceae. PeerJ, 7, e7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. Y. , Ye, J. W. , Wang, T. M. , Bao, L. , & Wang, H. F. (2020). Different processes shape the patterns of divergence in the nuclear and chloroplast genomes of a relict tree species in East Asia. Ecology and Evolution, 10(10), 4331–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich, M. , Lehwark, P. , Pellizzer, T. , Ulbricht‐Jones, E. S. , Fischer, A. , Bock, R. , & Greiner, S. (2017). GeSeq – Versatile and accurate annotation of organelle genomes. Nucleic Acids Research, 45(W1), W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonti‐Filippini, J. , Nevill, P. G. , Dixon, K. , & Small, I. (2017). What can we do with 1000 plastid genomes? The Plant Journal, 90(4), 808–818. [DOI] [PubMed] [Google Scholar]
- Tsui, H. P. (1987). A study on the system of Lindera . Acta Phytotaxonomica Sinica, 25(3), 161–171. [Google Scholar]
- Wilting, A. , Courtiol, A. , Christiansen, P. , Niedballa, J. , Scharf, A. K. , Orlando, L. , Balkenhol, N. , Hofer, H. , Kramer‐Schadt, S. , Fickel, J. , & Kitchener, A. C. (2015). Planning tiger recovery: Understanding intraspecific variation for effective conservation. Science Advances, 1(5), e1400175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, T. W. , Xu, Y. , Jin, L. , Liu, T. J. , Yan, H. F. , & Ge, X. J. (2020). Conflicting phylogenetic signals in plastomes of the tribe Laureae (Lauraceae). PeerJ, 8, e10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, T. W. , Yan, H. F. , & Ge, X. J. (2022). Plastid phylogenomics of tribe Perseeae (Lauraceae) yields insights into the evolution of east Asian subtropical evergreen broad‐leaved forests. BMC Plant Biology, 22(1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. H. , Liu, Q. , Hu, W. , Wang, T. , Xue, Q. , & Messing, J. (2015). Dynamics of chloroplast genomes in green plants. Genomics, 106(4), 221–231. [DOI] [PubMed] [Google Scholar]
- Xu, L. L. , Yu, R. M. , Lin, X. R. , Zhang, B. W. , Li, N. , Lin, K. , Zhang, D. Y. , & Bai, W. N. (2021). Different rates of pollen and seed gene flow cause branch‐length and geographic cytonuclear discordance within Asian butternuts. New Phytologist, 232(1), 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. , Sahu, S. K. , Yang, L. , Liu, Y. , Mu, W. , Liu, X. , Strube, M. L. , Liu, H. , & Zhong, B. (2022). Comparative analyses of 3,654 plastid genomes unravel insights into evolutionary dynamics and phylogenetic discordance of green plants. Frontiers in Plant Science, 13, 808156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution, 15(5), 568–573. [DOI] [PubMed] [Google Scholar]
- Ye, J. W. , Bai, W. N. , Bao, L. , Wang, T. M. , Wang, H. F. , & Ge, J. P. (2017). Sharp genetic discontinuity in the aridity‐sensitive Lindera obtusiloba (Lauraceae): Solid evidence supporting the tertiary floral subdivision in East Asia. Journal of Biogeography, 44(9), 2082–2095. [Google Scholar]
- Zekun, L. , & Haixia, C. (2012). GC‐MS analysis of essential oil from the bark of Lindera obtusiloba . Chemistry of Natural Compounds, 48(4), 696–697. [Google Scholar]
- Zhang, R. S. , Yang, J. , Hu, H. L. , Xia, R. X. , Li, Y. P. , Su, J. F. , Li, Q. , Liu, Y. Q. , & Qin, L. (2020). A high level of chloroplast genome sequence variability in the sawtooth oak Quercus acutissima . International Journal of Biological Macromolecules, 152, 340–348. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. H. , Wang, I. J. , Comes, H. P. , Peng, H. , & Qiu, Y. X. (2016). Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a tertiary relict species, Emmenopterys henryi (Rubiaceae). Scientific Reports, 6, 24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. J. , Liang, T. T. , & Zang, D. K. (2014). Analysis on community composition and structure of Lindera obtusiloba in Laoshan. Journal of Agriculture, 4(4), 60–63. [Google Scholar]
- Zhao, M. L. , Song, Y. , Ni, J. , Yao, X. , Tan, Y. H. , & Xu, Z. F. (2018). Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae). Scientific Reports, 8(1), 8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S. , Poczai, P. , Hyvönen, J. , Tang, J. , & Amiryousefi, A. (2020). Chloroplot: An online program for the versatile plotting of organelle genomes. Frontiers in Genetics, 11, 576124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.
Appendix S2.
Appendix S3.
Appendix S4.
Data Availability Statement
The Lindera obtusiloba plastomes generated in this study are available in the NCBI GenBank repository with accession numbers ON520645 to ON520654. The Supplementary data to this article can be found online at https://10.1002/ece3.11119.


