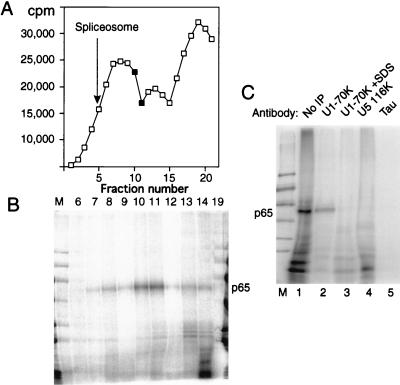
FIG. 7.
p65 cross-linking in U1 snRNP-containing complexes. Full-length labeled GC+DX RNA was incubated in nuclear extract for 15 min and then fractionated on a 10 to 30% glycerol gradient. (A) Profile of Cerenkov counts across the glycerol gradient. Fractions are numbered from 1 (heaviest) to 20 (lightest). Fully assembled spliceosomes, as indicated by the presence of splicing intermediates, were shown to peak in fraction 5 in control gradient fractionations. The complex that forms at time zero peaks at fraction 13. (B) MB cross-linking of gradient fractions from panel A. Cross-linked p65 was reproducibly maximal in fractions 10 and 11. Note that the lane containing fraction 19 has been contaminated by spillover of size markers. Nevertheless, this fraction can still be seen to be essentially free of cross-linked p65. (C) Labeled GC+DX XhoI RNA was incubated in nuclear extract and then MB cross-linked and electrophoresed by SDS-PAGE after no further treatment (lane 1) or after immunoprecipitation with monoclonal antibody H111 against U1 70K protein (lanes 2 and 3), antiserum against U5 116K protein (lane 4), or a control monoclonal antibody against tau protein (lane 5). In lane 3, the cross-linked sample was first pretreated with 0.8% SDS to disrupt U1 snRNPs before dilution into NETS buffer and immunoprecipitation with H111. p65 was immunoprecipitated by the U1 70K antibodies, but only if U1 snRNP remained intact (lane 2).