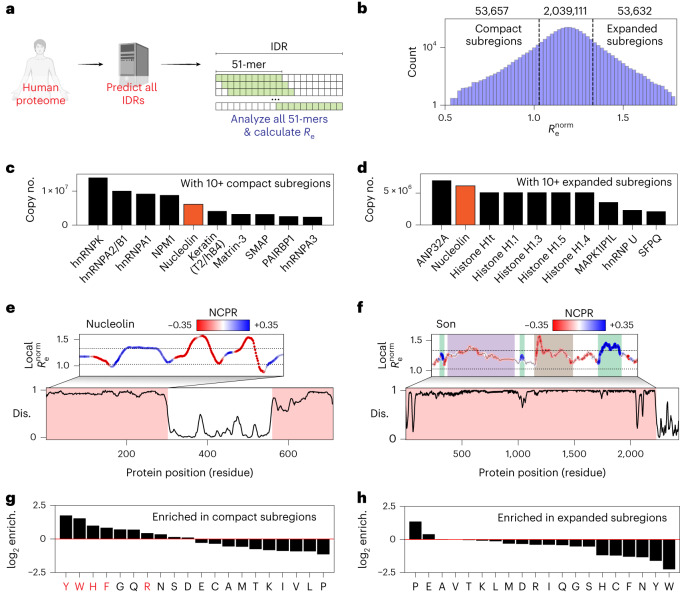
Fig. 5. Local analysis of disordered protein subregions reveals sequence-dependent expansion and compaction.
a, Graphical summary illustrating the sliding-window subregion analysis presented in this figure. b, Distribution of the normalized end-to-end distance obtained from all 51-residue subfragments within IDRs in the human proteome (note y axis is a log scale). c, Proteins with ten or more compact subregions ordered by copy number in HeLa cell lines. d, Proteins with ten or more expanded subregions ordered by copy number in HeLa cell lines. e, Linear analysis of the first IDR in the highly abundant nucleolar protein nucleolin. Dashed lines represent the same dashed lines as shown in b. Nucleolin possesses both compact and expanded subregions (c and d), as highlighted in this linear analysis (UniProt P19338). f, Linear analysis of local subregions in the 2,227-residue IDR from the nuclear speckle protein Son. Dashed lines represent the same dashed lines as shown in b. Son possesses several conformationally distinct subregions, as highlighted (UniProt P18583). g, log2 fold enrichment for amino acids found in compact subregions. Residues implicated in RNA binding are highlighted in red. h, log2 fold enrichment for amino acids found in expanded subregions.