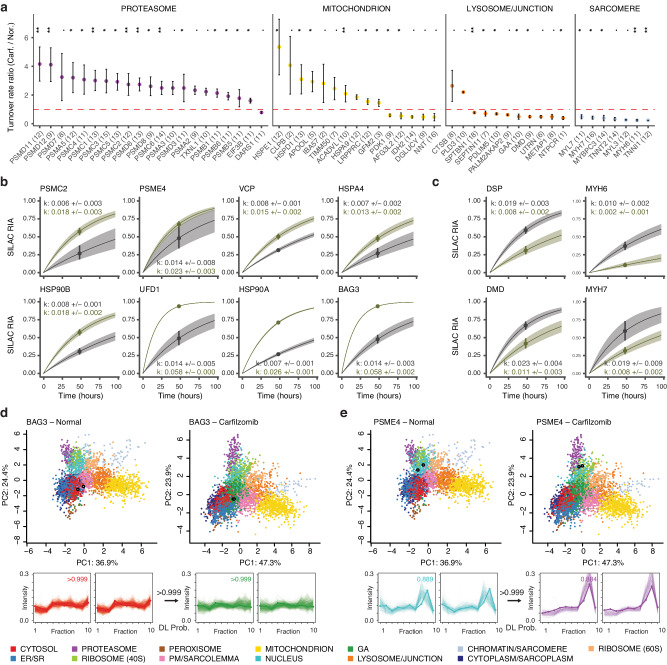
Fig. 7. Proteostatic pathways and lesions in carfilzomib mediated cardiotoxicity in iPSC-CMs.
a Changes in protein turnover rates between carfilzomib vs. normal iPSC-CMs across selected cellular compartments; ∙: P < 0.1; *: <0.05; ** <0.01; Mann-Whitney test (two-sided) with Benjamini-Hochberg multiple-testing correction. Dashed line: 1:1 ratio; point: median ratio; range: median ± MAD/median of ratios; n=number of peptide observations (parenthesis) over 2 biologically independent samples per gorup. Source data are provided as a Source Data file. b Kinetic curve representations of proteins with accelerated temporal kinetics in carfilzomib, including PSMC2 which corresponds to the ratio in panel A, as well as additional ERAD proteins and chaperones; gray: normal iPSC-CM; green: carfilzomib. Point: best-fit k; line: the first-order kinetic curve of k; bands: fitting s.e. c Kinetic curve representations of slowdown of protein kinetics in DSP, DMD, MYH6, and MYH7, corresponding to the ratios in panel a. Point: best-fit k; line: first-order kinetic curve of k; bands: fitting s.e. d, e Spatial map (PC1 vs. PC2) and ultracentrifugation fraction profiles of d. BAG3 and e. PSME4 in normal and carfilzomib-treated human iPSC-CM, showing a likely differential localisation in conjunction with kinetics changes. White-filled circles: light and heavy BAG3 or PSME4 in each plot. The kinetic curves of BAG3 and PSME4 are in panel b. Numbers at arrows correspond to BANDLE differential localization (DL) probability.