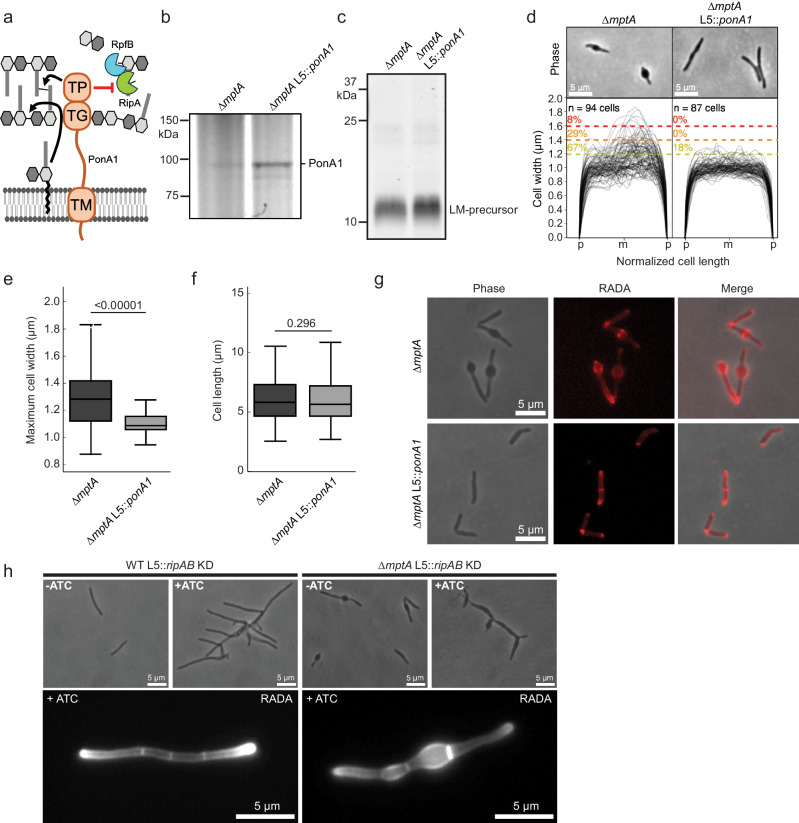
Fig. 4. PonA1 rescues the morphological defects of ∆mptA.
a The domain structure of PonA1 and the corresponding cell wall substrates and interacting partners. The C-terminal domain of PonA1 interacts with RipA to suppress the activity of the peptidoglycan hydrolase complex formed by RipA and RpfB. TG, transglycosylase domain; TP, transpeptidase domain; TM, transmembrane domain. Cartoon made with Microsoft PowerPoint. b SDS-PAGE of ∆mptA and ∆mptA L5::ponA1 cell lysates in which PonA1 is stained with the fluorescent penicillin analog bocillin. Gel is representative of two independent experiments with similar results. c SDS-PAGE of lipoglycans extracted from ∆mptA and ∆mptA L5::ponA1, visualized by glycan staining, showing that ponA1 overexpression does not restore LM/LAM biosynthesis. Gel is representative of two independent experiments with similar results. d Phase contrast micrographs and cell width profiles of ∆mptA cells and ∆mptA cells overproducing PonA1 (∆mptA L5::ponA1). Cell width profiles were graphed as described in Fig. 2. “p” and “m” indicate cell poles and mid-cell, respectively. e, f Boxplots comparing the distribution of maximum cell widths (e) and cell lengths (f) between ∆mptA and ∆mptA L5::ponA1. n = 94, 87 cells for ∆mptA and ∆mptA L5::ponA1, respectively. The boxplots are drawn as described in Fig. 1. Statistical significance was determined using the one-tailed student’s T-test. In panel (e), P = 3.58 × 10−13. g Phase contrast and fluorescence micrographs of RADA-labeled ∆mptA and ∆mptA L5::ponA1 cells. Microscope images are representative of two independent experiments with similar results. h Top: phase contrast micrographs of ATC-inducible ripAB CRISPRi knockdown strains constructed in the WT and ∆mptA genetic backgrounds. ripAB KD was induced for 20 h. Bottom: fluorescent micrographs of ATC-inducible ripAB KD strains stained with RADA. ripAB KD was induced for 8 h. Microscope images are representative of two independent experiments with similar results. Source data for panels b–f are provided in the Source Data file. Microscopy image data for panels d–h are available at https://github.com/IanLairdSparks/Sparks_2023.