Figure 3: Microbial genes metatranscriptomically implicated in generation of N-acetyl 5-ASA cluster into thiolase and acyl CoA N-acyltransferase superfamilies.
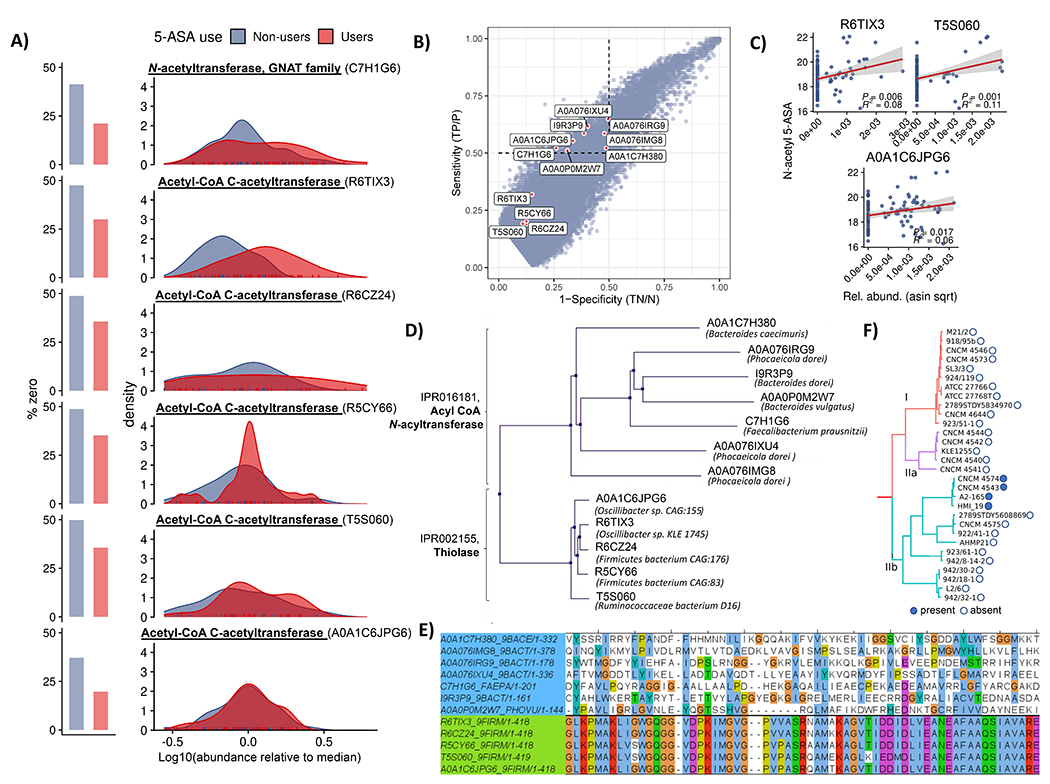
(A) MTX abundance distributions for six putative 5-ASA-acetylating gene families in 5-ASA users vs non-users. Multivariate linear mixed effects models adjusted for DNA copy number (27) identified two significantly overexpressed gene clusters with putative acetyltransferase function in 5-ASA users vs. non-users: 1) a GNAT family N-acetyltransferase (UniRef90 ID: C7H1G6) and 2) an acetyl-CoA acetyltransferase (UniRef90 ID: R6TIX3) (FDR q 0.24 and 0.14, respectively). Searching for any additional sequences with at least 80% full-length sequence similarity yielded three additional hits, all from the first acetyl-CoA acetyltransferase, each nominally enriched in 5-ASA users compared to non-users. (B) Specificity and sensitivity for microbial transcripts with respect to presence/absence of fecal N-acetyl 5-ASA across samples from 5-ASA users identifies seven additional putative 5-ASA acetylating gene families. In our second criteria, we estimated sensitivity and specificity for how each metatranscriptomic gene cluster (presence/absence) detected dichotomized N-acetyl 5-ASA (high/low). Using a 50% cutoff (hashed line) for each test characteristic revealed an additional 7 putative acetyltransferase gene clusters. Shown outside of these bounds are results from the first criteria, highlighting high degrees of specificity, but lower sensitivity. (C) MTX relative abundances for three of these MBX-based candidates identified in the second criteria also correlated with fecal N-acetyl 5-ASA levels (R2 and p values inset). Error bands represent 95% confidence intervals. (D) Pooled metatranscriptomic families from parts A and B proved to cluster into two protein superfamilies – thiolase and acyl-CoA N-acyltransferases – which are carried primarily by Bacteroides and Firmicutes phylum members, respectively. (E). Multiple sequence alignment of these twelve candidate enzymes shows highly conserved sequences among the thiolase enzymes (green) but greater diversity among acyltransferases (light blue). (F) In genomes from isolate strains, the only acyl-CoA N-acyltransferase gene carried by a Firmicutes member was in a subset of F. prausnitzii strains. When hierarchically clustered according to a prior schema33 (repurposed with permission), these appeared to originate from only one of the clade’s major phylogroups, suggestive of acquisition via a horizontal transfer event.
