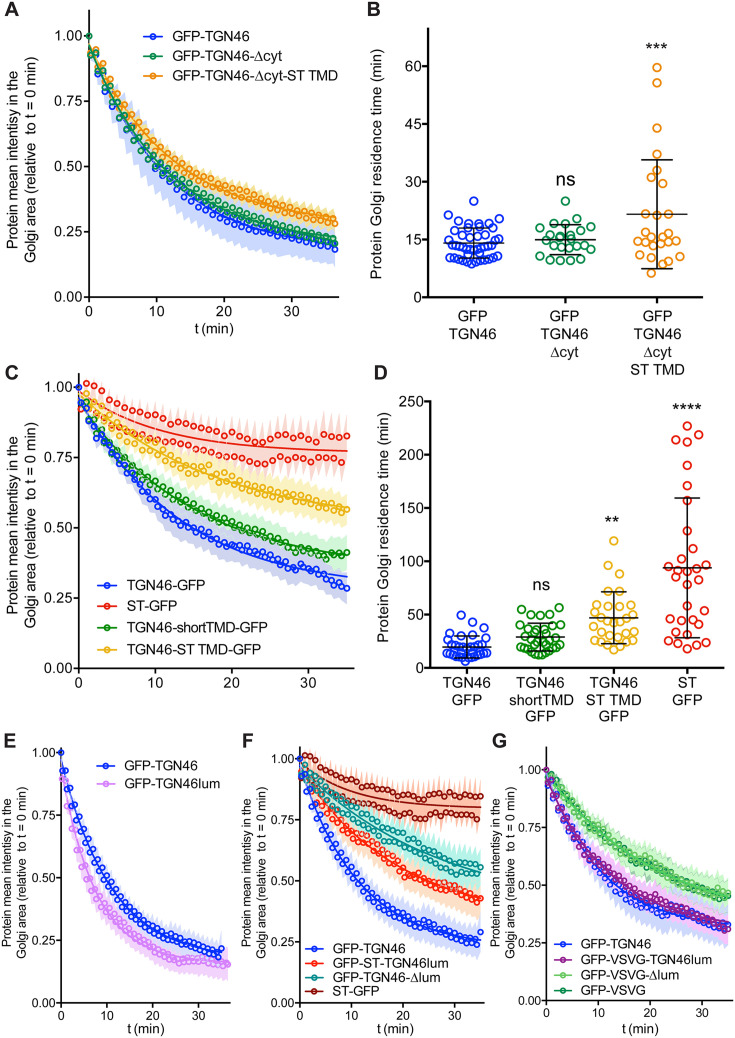
Figure 3. Fluorescence loss in photobleaching (FLIP) experiments monitor Golgi residence times of different proteins.
(A, C, E–G) Relative fluorescence intensity average time trace (mean ± standard error of the mean [s.e.m.]) of FLIP experiments for the indicated proteins. Symbols correspond to actual measurements, solid lines to the fitted exponential decays. (B, D) Residence time in the perinuclear area measured as the half time of the FLIP curves. Results are from 7 to 12 cells from each of n = 3 independent experiments (individual values shown, with mean ± stdev; ns, p > 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001).