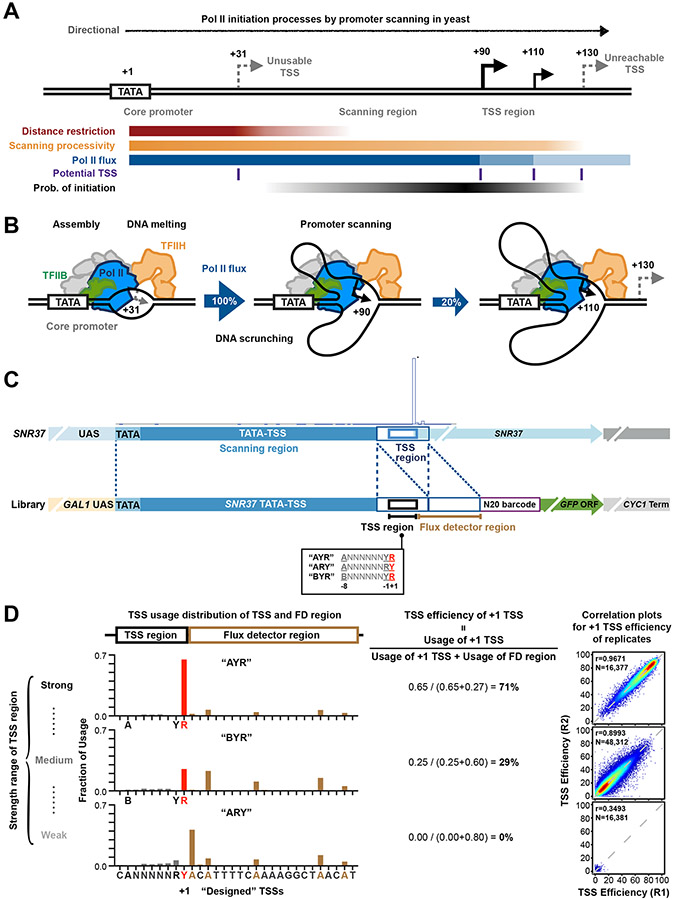
Figure 1. A high-throughput system for studying transcription TSS selection.
(A and B) Pol II initiation in yeast proceeds by promoter scanning. Yeast Pol II initiation usually occurs at multiple TSSs ~40 to 120 bp downstream of a core promoter comprising the PIC assembly position (e.g. a TATA element)). After PIC assembly upstream, scanning will proceed towards positions where TSS selection occurs (TSS region). Initiation across promoter positions is also controlled by multiple architectural features shown in A. These include the inhibition of initiation near a core promoter that diminishes downstream (“distance restriction”), biochemical restrictions on how far scanning can proceed downstream (“scanning processivity”), and “Pol II flux”, which represents the decrease in amount of scanning Pol II due to conversion of scanning Pol II initiating. (C) Construction of promoter libraries examining TSS sequence context. Top panel shows schematic of the SNR37 promoter and its TSS distribution based on TSS-seq 21. Bottom panel shows schematic of the Pol II MASTER libraries. A duplication of the SNR37 TSS region was inserted before native TSS region, and the −8 to +1 positions relative to native SNR37 +1 TSS (black box) were replaced by a 9 nt highly randomized region. The downstream SNR37 TSS region functions as a “Flux Detector” (FD) to capture Pol II that fails to initiate within the randomized region and allow measurement of initiation efficiency for upstream positions. A barcode (purple box) allows RNA products to be assigned to promoter variants. Other features (GAL1 UAS, GFP ORF, CYC1 terminator) support regulation and stabilization of RNAs. (D) TSS usage distributions at TSS and FD regions for promoter variant “AYR”, “BYR”, and “ARY” libraries are shown (left). TSS usages from designed +1 TSS and positions upstream are red/grey, respectively. TSS usage from the FD region is in brown. The definition of “TSS efficiency” and overall TSS efficiencies for aggregate +1 TSSs for different libraries are shown (middle). Example correlation plots of TSS efficiency calculations for +1 TSSs from individual promoter variants in Pol II MASTER libraries between representative biological replicates are shown (right) with Pearson r and N of variants.