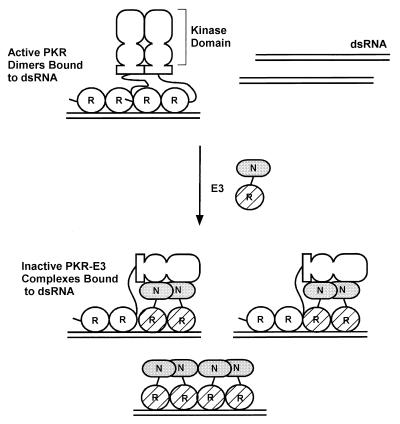
FIG. 11.
Hypothetical model for inhibition of PKR function by E3 through the formation of inactive heteromeric complexes. PKR is shown schematically with its two DRBMs (R) connected by a linker to the dimerization domain located between residues 244 and 296 (shown as a rectangle) and the N-terminal and C-terminal lobes of the kinase domain (depicted as two ovals). E3 is depicted with its single DRBM (R) hatched and the N-terminal domain (N) shaded. The active form of PKR is depicted as a dimer bound to dsRNA (28, 31, 40), with dimerization mediated by interactions involving the N-terminal region containing the DRBMs (12, 37, 48, 50), the kinase domain (37, 40), and the region from residues 244 to 296 (48) and by binding to the same dsRNA molecule (12, 37, 51). E3 is shown inhibiting PKR by forming inactive heterocomplexes, disrupting PKR homodimers. In addition, the N-terminal domain of E3 is shown interacting with the kinase domain of PKR, interfering with some aspect of kinase function. Binding to dsRNA by E3 greatly contributes to the stability of the PKR-E3 complex. E3 can also inhibit kinase activation by sequestering dsRNA molecules. See text for details.