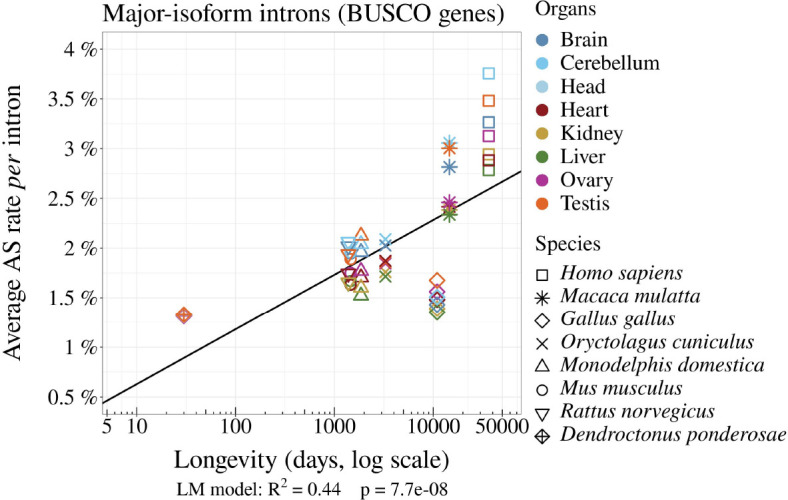
Figure 3. The rate of alternative splicing correlates with life history traits across metazoans.
(A) Relationship between the per intron average AS rate of an organism and its longevity (days, log scale). (B) Variation in average AS rate across seven organs (brain, cerebellum, heart, liver, kidney, testis, and ovary) among seven vertebrate species (RNA-seq data from Cardoso-Moreira et al., 2019). AS rates are computed on major-isoform introns from BUSCO genes (Materials and methods).
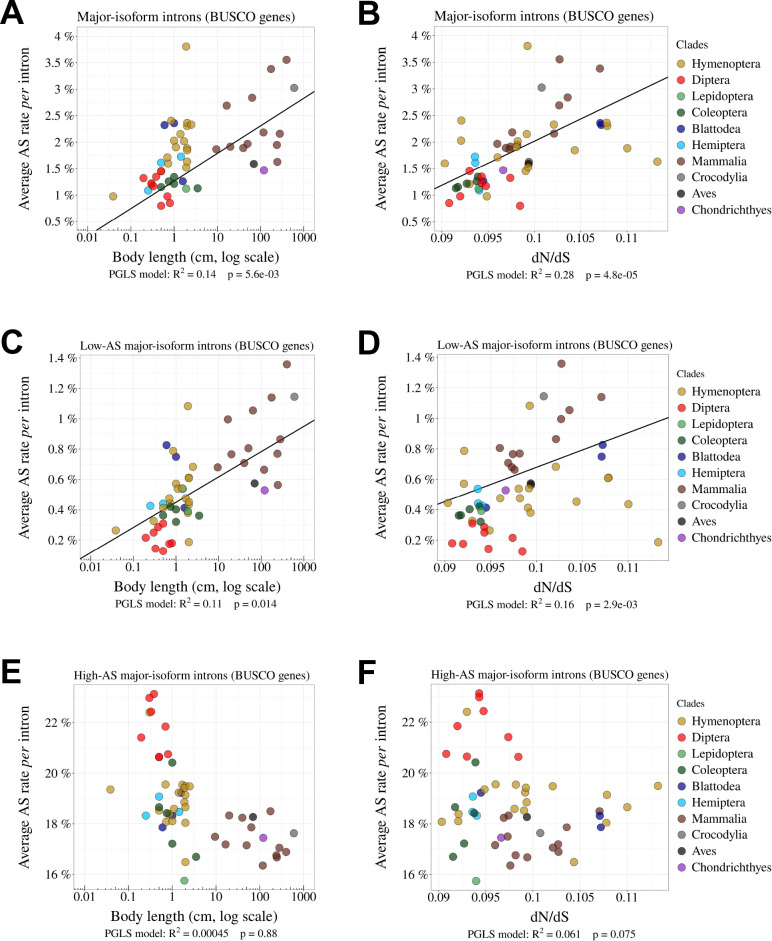
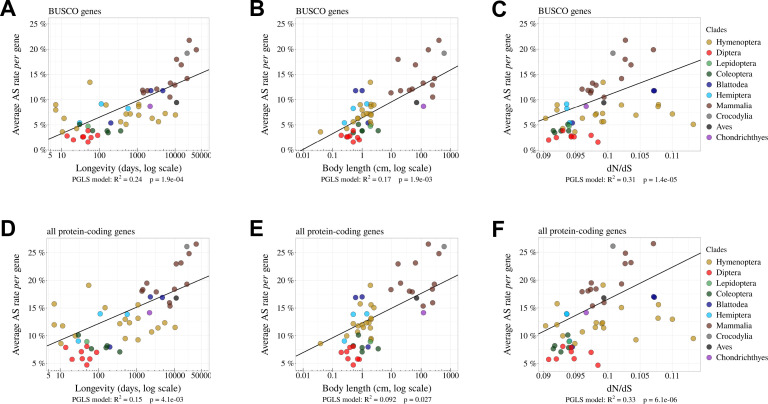
Figure 3—figure supplement 1. Relationship between AS rates and other Ne proxies.
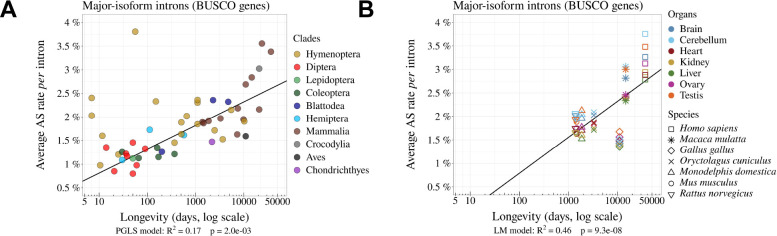
Figure 3—figure supplement 2. The rate of alternative splicing correlates with life history traits in both vertebrates and insects.
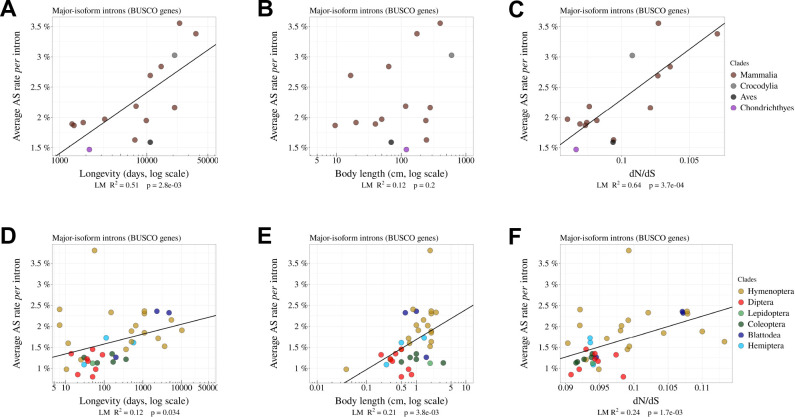
Figure 3—figure supplement 3. The variation in AS rates between species is not explained by organ differences.