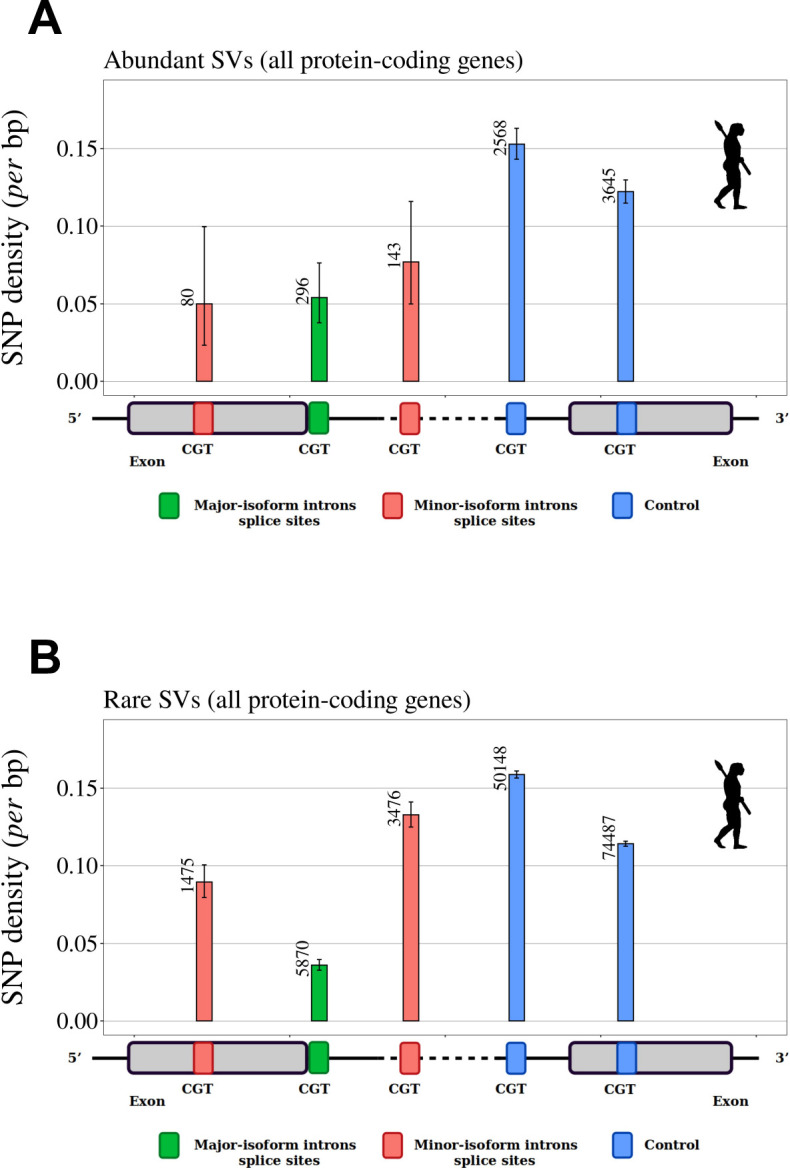
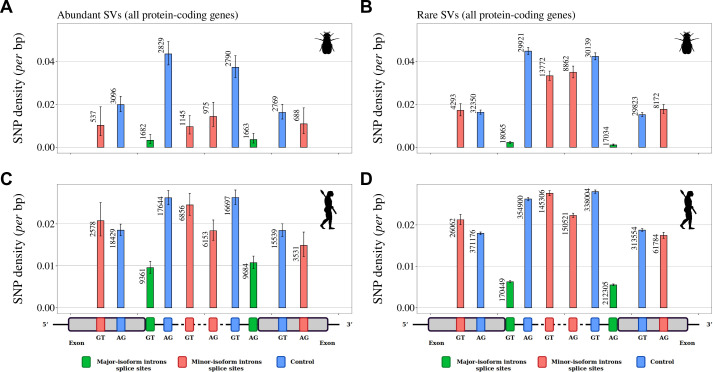
Figure 5. Variation in selective constraints on alternative splice signals from rare and abundant SVs.
For each minor-isoform intron sharing one boundary with a major-isoform intron, we measured the SNP density at its minor splice site (red), and at the corresponding major splice site (green). We distinguished minor splice sites that are located in an exon or in an intron of the major isoform. As a control (blue), we selected AG or GT dinucleotides that are unlikely to correspond to alternative splice sites, namely: AG dinucleotides located toward the end of the upstream exon or the beginning of the intron (unlikely to correspond to a genuine acceptor site), and GT dinucleotides located toward the beginning of the downstream exon or the end of the intron (unlikely to correspond to a donor site). To increase the sample size, we analyzed data from all annotated protein-coding genes (and not only the BUSCO gene set). The number of sites studied is shown at the top of each bar. Error bars represent the 95% confidence interval of the proportion of polymorphic sites (proportion test). (A,B) SNP density in Drosophila melanogaster (polymorphism data from 205 inbred lines derived from natural populations, N=3,963,397 SNPs Huang et al., 2014; Mackay et al., 2012). (C,D) SNP density in Homo sapiens (polymorphism data from 2504 individuals, N=80,868,061 SNPs Auton et al., 2015). We excluded dinucleotides affected by CpG hypermutability (Materials and methods, see Figure 5—figure supplement 1 for CpG sites). (A,C) Abundant SVs (MIRA > 5%). B,D: Rare SVs (MIRA ≤ 5%).
Figure 5—figure supplement 1. SNP density in human splice signals, for dinucleotides affected by CpG hypermutability.