FIGURE 1.
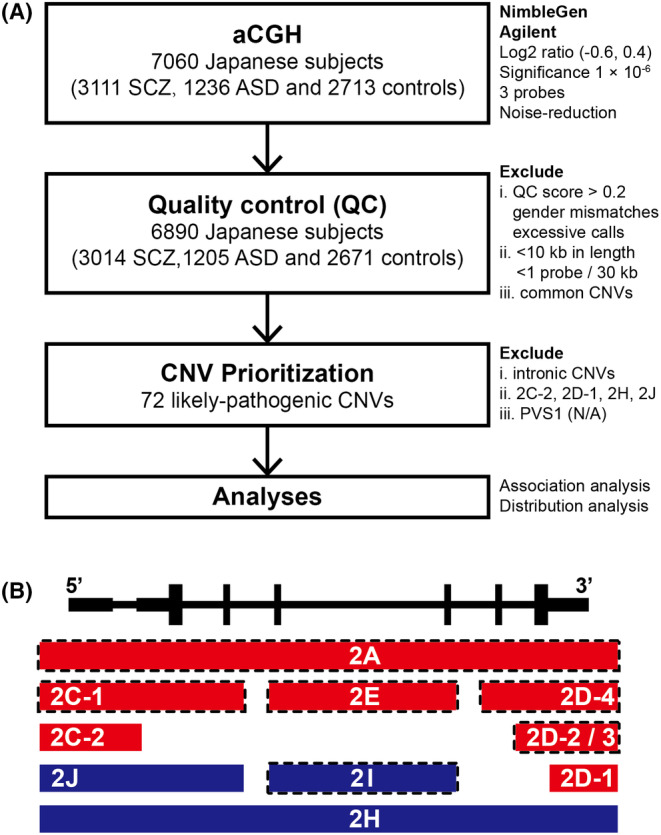
Workflow and classifications of CNVs in the present study. (A) Workflow of the present study. (B) Schematic illustration of classifications based on overlapping patterns following the guidelines for the interpretation of CNVs provided by ACMG and ClinGen. Red bars and dark blue bars represent deletions and duplications, respectively. Bars framed by the dotted line represent classifications prioritized as LP‐CNVs. 2A: Deletions completely overlapping an established HI gene/genomic region; 2C‐1: Deletions partially overlapping with the 5′ end (3′ end not involved) and the coding sequence is involved; 2C‐2: Deletions partially overlapping with the 5′ end (3′ end not involved) and only the 5’ UTR is involved; 2D‐1: Deletions partially overlapping with the 3′ end (5′ end not involved) and only the 3′ untranslated region is involved; 2D‐2/3: Deletions partially overlapping with the 3′ end (5′ end not involved) and only the last exon is involved; 2D‐4: Deletions partially overlapping with the 3′ end (5′ end not involved) and includes other exons in addition to the last exon. Nonsense‐mediated decay is expected to occur; 2E: Deletions with both breakpoints within the same gene; 2H: Duplications fully containing an HI gene; 2I: Duplications with both breakpoints within the same gene; 2 J: Duplications with one breakpoint within the established HI gene. ACMG, American College of Medical Genetics and Genomics; ClinGen, Clinical Genome Resource; HI, haploinsufficient; LP‐CNV, likely pathogenic copy number variation.
