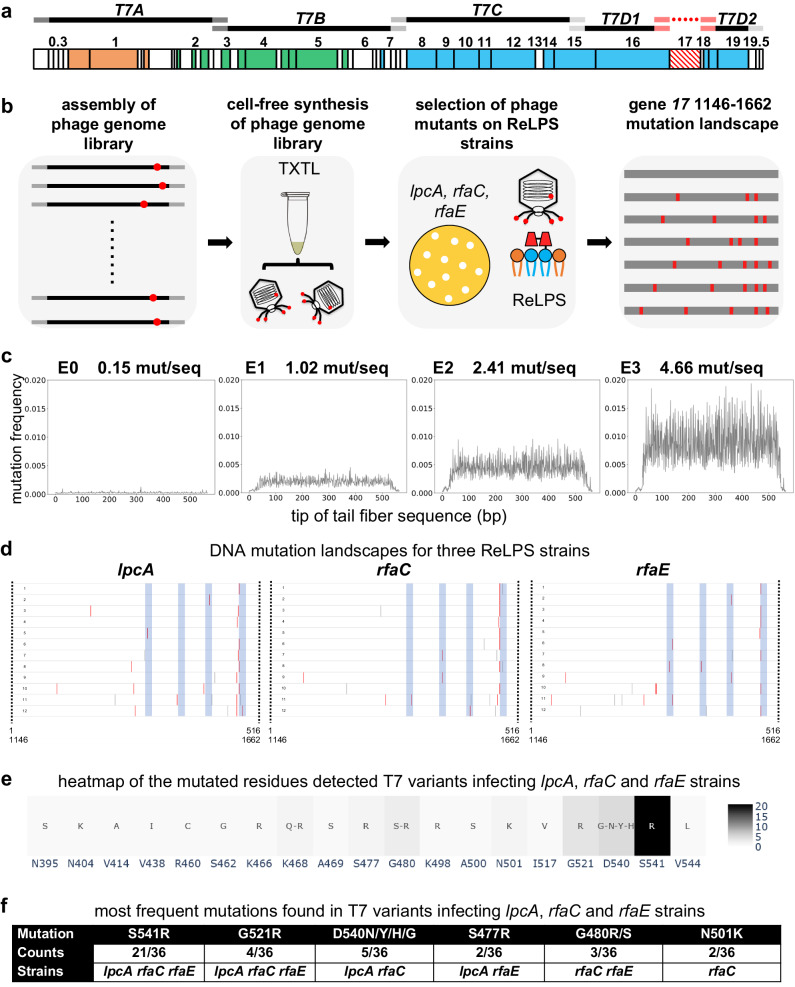
Fig. 5. Using PHEIGES to engineer T7 phages that infect E. coli strains with any type of rough LPS.
a The T7 genome is assembled using five T7 WT gene parts (T7A, T7B, T7C, T7D1, T7D2) and the fragment 1146–1662 of the tail fiber gene 17 obtained by mutagenic PCR. b PHEIGES assembly workflow for T7 LPS mutant libraries. T7 genomes with E0, E1, E2 and E3 were separately assembled and expressed in TXTL (cell-free transcription-translation). TXTL reaction containing the phage libraries were directly spotted on E. coli strain harboring ReLPS. Twelve T7 variants were selected, and their tail fiber sequenced for each of the three E. coli mutant strains lpcA, rfaC and rfaE (all ReLPS), 36 phages total. E. coli B strain has a type RbLPS. c The mutation frequency of the four mutated tail fiber DNA fragments was determined by NGS. The graphs are indexed from 1 to 516 which corresponds to the bp 1146–1662 of gene 17. Source data are provided as a Source Data file. d The tail fibers of the selected phages were sequenced to establish the mutation landscape, especially the T7-ReLPS. The blue zones show the external loops of the tail fiber tip. Gray bars are silent mutations, red bars are non-silent mutations. 6 plaques were picked from T7-E1 (6 first from the top in Fig. 5d, labeled 1–6), 3 plaques from T7-E2 (three following in Fig. 5d, labeled 7–9), and 3 plaques from T7-E3 (last three in Fig. 5d, labeled 10–12) from rfaC, lpcA, and rfaE. Source data are provided as a Source Data file. e This heatmap compiles all the mutations of the 3 × 12 phages. the native amino acid is indicated below every mutation and the color scale indicates how many times a mutation is found in the 36 variants. f This table reports the most frequent amino acid mutations among the 36 T7 variants that lead to ReLPS infection as a single mutation.