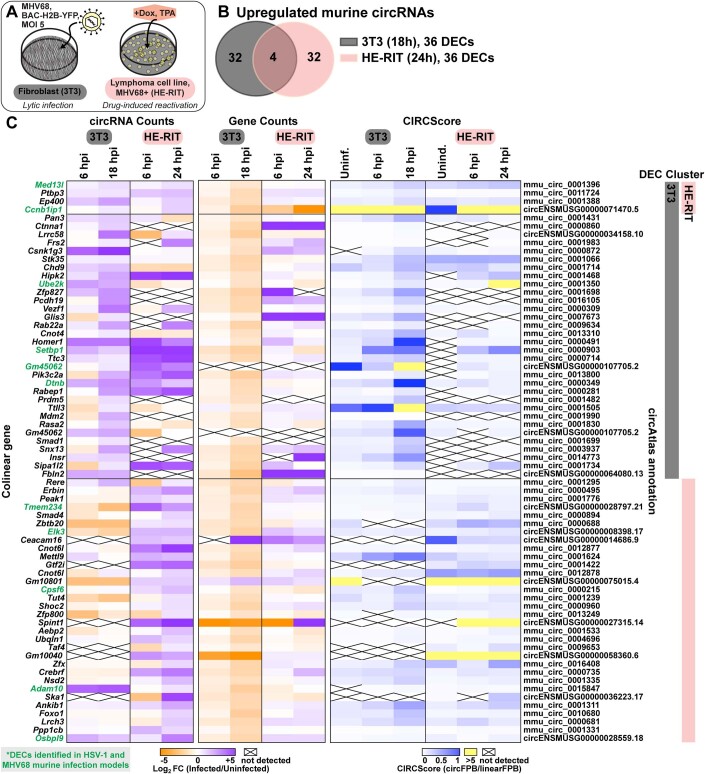
Figure EV2. Murine circRNAs upregulated in MHV68 infection models.
(A) Infographic for murine MHV68 infection models used in this study. (B) Overlap of upregulated murine circRNA detected by bulk RNA-Seq from MHV68 de novo infection (NIH 3T3, n = 2) and drug-induced reactivation (HE-RIT, n = 2). (C) Heatmaps for upregulated murine circRNAs, plotted as circRNA counts (log2FC), Gene counts (log2FC), or CIRCScore (circFPB/linearFPB). Bulk RNA-Seq data was normalized to ERCC spike-in controls. Log2FC is relative a paired uninfected or uninduced control. Differential expression p-values were calculated using RankProd non-parametric permutation tests. DECs were those with raw back splice junction (BSJ) count across the sample set >10, log2FC > 0.5, and p-value < 0.05. Heatmap values are the average of biological replicates.