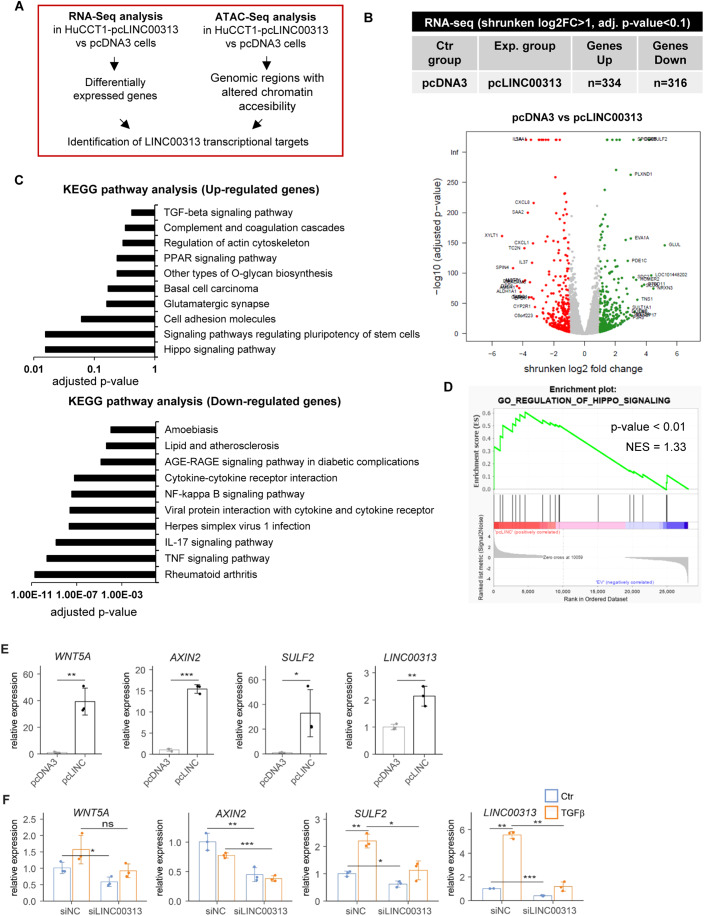
Figure 3. LINC00313 regulates genes involved in the Wnt pathway.
(A) Experimental approach to identify LINC00313 transcriptional targets. (B) Number of differentially expressed genes in pcLINC00313 versus pcDNA3 HuCCT1 cells (shrunken log2FC > 1, adjusted p-value < 0.1). Volcano plot shows differentially expressed genes (green colour: upregulated genes, red colour: downregulated genes). (C) KEGG pathway analysis of up or downregulated genes upon LINC00313 over-expression. (D) Example of GSEA of differentially expressed genes in response to LINC00313 over-expression. Shown is the enrichment of a Hippo signalling signature in the gene expression of cells overexpressing LINC00313. (E) WNT5A, AXIN2 and SULF2 mRNA levels in HuCCT1 expressing LINC00313 (pcLINC) or empty vector (pcDNA3). (F) WNT5A, AXIN2 and SULF2 expression in HuCCT1 transfected with siLINC00313 and stimulated with TGFβ1 or BSA/HCl for 16 h. Data information: In panel (B) statistically significant differential expressed genes were identified using DEseq2. In panel (C) adjusted p-value was calculated using the Benjamini-Hockberg method for correction for multiple hypotheses testing. In panel (D) GSEA results are displayed as a normalized enrichment score (NES) and presented as an enrichment plot. The Kolmogorov-Smirnov test was used for statistical analysis of GSEA. In (E,F) data are presented as mean ± SD (n = 3 biological replicates). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test). Source data are available online for this figure.