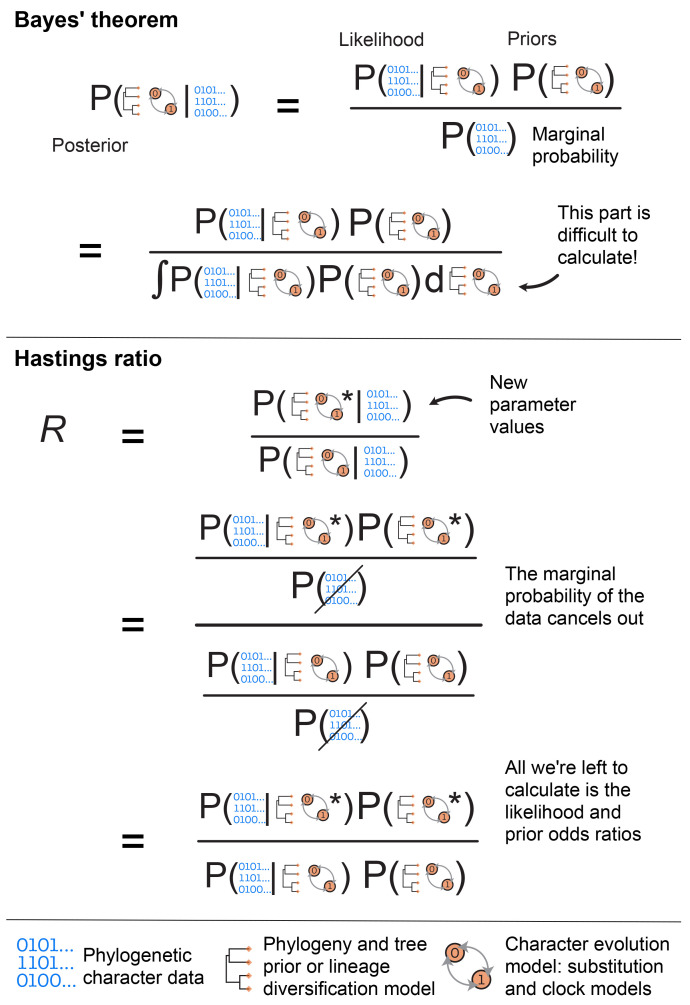
Figure 1. Schematic representation of Bayes’ theorem and the Hastings ratio, used in Bayesian inference.
The top panel shows Bayes’ theorem and the relationship between the posterior, likelihood, priors and the marginal probability of the data. The second line of this panel shows an alternative way of writing the marginal probability, which illustrates more explicitly why the marginal probability is difficult to calculate. During MCMC we sample new parameter values (indicated by * in the figure) at each step and compare their posterior probability to the previous set of values using the Hastings or posterior odds ratio, R. The middle panel shows the Hastings ratio, and illustrates that since the marginal probability cancels out, we avoid having to calculate it during MCMC. The bottom panel provides a legend for the main data and model components used in Bayesian phylogenetic inference. The observed data used to infer phylogenies typically comprise discrete morphological characters (shown here) or molecular sequences. During inference the tree topology and branch lengths are sampled from a prior distribution that depends on the type of inference − in particular, if the tree is dated we typically use lineage diversification models, such as birth-death processes. The character evolution model will always include the substitution process and will additionally include the clock model if we aim to infer a dated phylogeny.