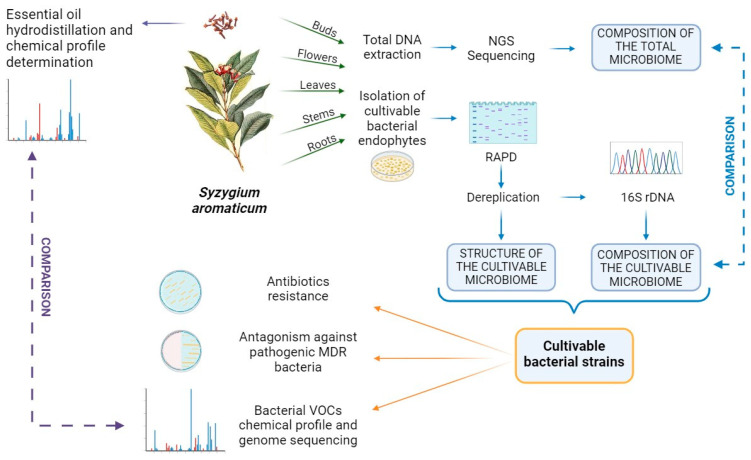
Figure 4.
Experimental workflow for the molecular and phenotypic characterization of bacterial endophytes associated with S. aromaticum. The essential oil is obtained from the buds (and/or the leaves) of the plant. After surface sterilization, small amounts of each anatomical part are used for both total DNA extraction and cultivable bacterial endophytes’ isolation. The DNA is sequenced to obtain the composition of the total (cultivable and non-cultivable) microbiota, while the culturable endophytic strains are subjected to both molecular and phenotypic analyses. Random Amplified Polymorphic DNA (RAPD) analysis and the 16S rRNA gene sequencing provide information on the structure and composition of each community, highlighting the degree of biodiversity and the possible microbiome divergences among different microenvironments. The collection of cultivable strains allows the phenotypic characterization of endophytes. It is possible to explore the levels of antibiotic resistance of each community and the ability of endophytes to interfere with the growth of MDR pathogens through the production of antibacterial molecules, mimicking the antimicrobial potential of the essential oil. The characterization of the volatile organic compounds (VOCs) produced by the strain could shed light on endophytes’ potential to produce similar, if not the same antibacterial VOCs and so contribute to the plant’s essential oil composition. (Created with BioRender.com, accessed on 8 January 2024).