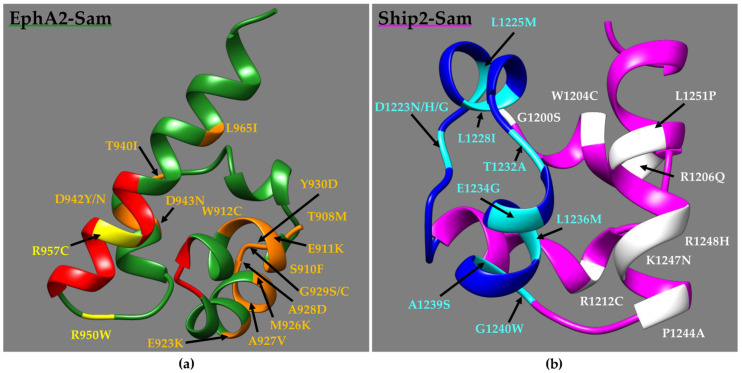
Figure 3.
(a) NMR structure of EphA2–Sam (dark green) (first conformer -residues T908–V972-, pdb entry 2E8N without flexible N- and C-terminal tails) in a ribbon representation. The EH interface (residues I916–M918 and P952–Y960) is highlighted in red, while diverse mutations are coloured in orange and yellow if positioned far away from or inside/close to the EH, respectively. Mutations V904G and R907C/S are not shown as located in the N-terminal flexible tail. (b) NMR Structure of Ship2–Sam (magenta) (first conformer -residues G1200–K1258-, pdb entry 2K4P [17] after removal of the flexible N-tail) in a ribbon representation. The ML interface (residues H1239–E1238) is coloured in blue; mutations are highlighted in white and cyan if positioned far away from or inside/close to the ML, respectively. Mutation E1198K is not shown as included in the N-terminal disordered tail.