Figure 7:
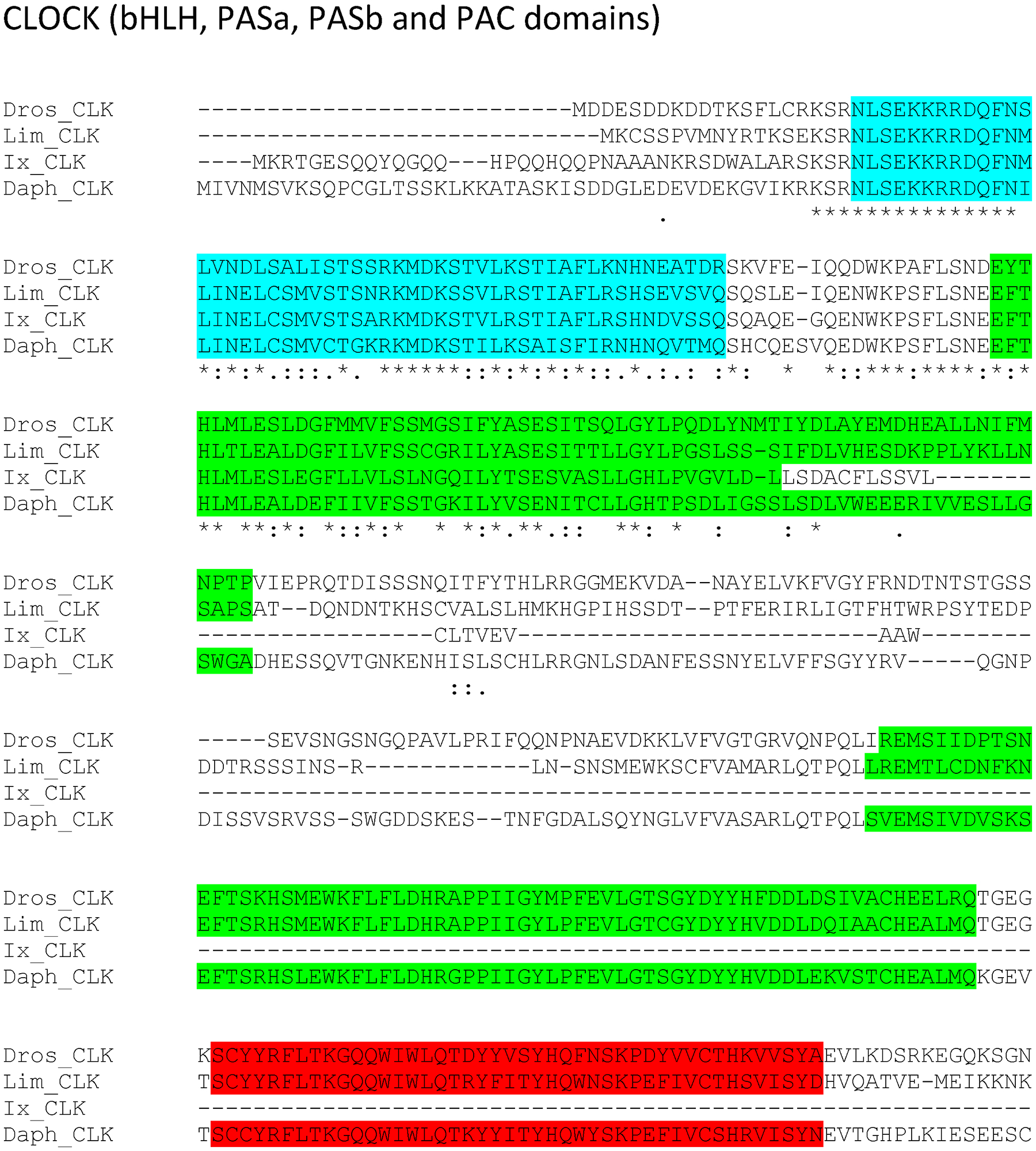
bHLH, PASa, PASb and PAC domains region of putative L. polyphemus CLOCK (CLK) protein. Alignment of L. polyphemus (Lim) CLK to Daphnia pulex (Daph) CLK, D. melanogaster(Dros) CLK, Ixodes scapularis (Ix) CLK using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, Light green shows PAS domain, Red shows PAC domain. For full alignment see Supplemental Figure 3.
