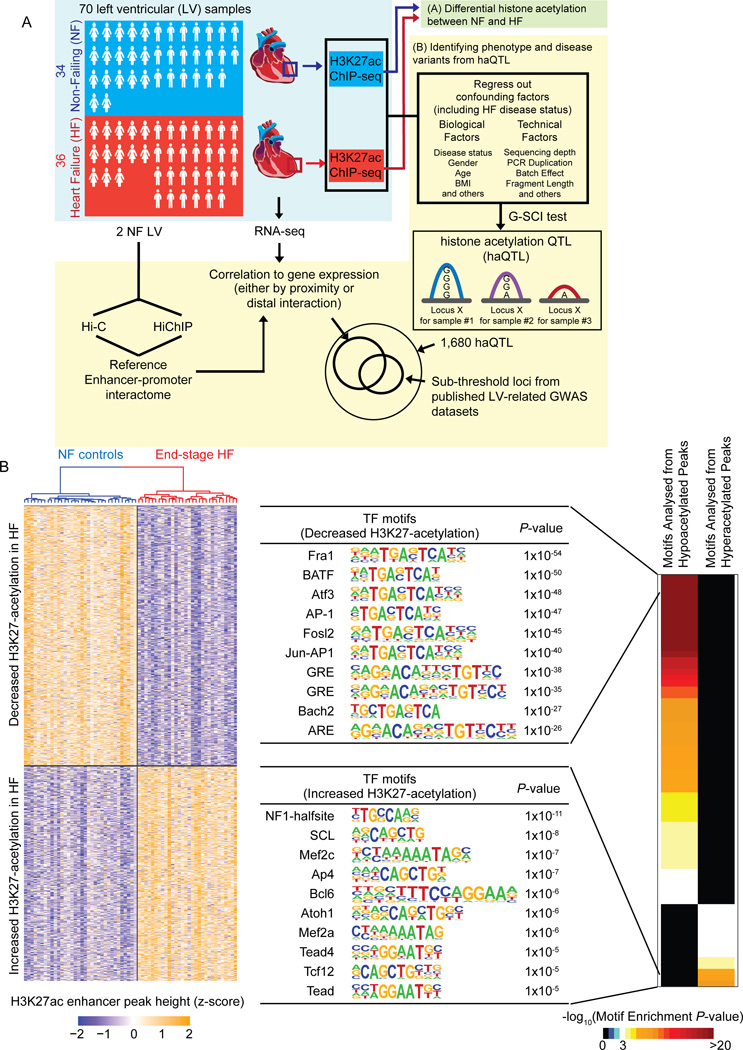
Figure 1. Schematic and differential H3K27-acetylation between non-failing control (NF) and failing (HF) hearts.
(A) Schematic for the experimental workflow. H3K27ac ChIP-seq was performed on 70 human left ventricles (LV). The first stage of the analysis involved (A), a direct analysis for differential histone acetylation between NF and HF. In the second stage (B), all ChIP-seq peaks were collected, and regressed for factors including HF disease status. The G-SCI test was performed to call out haQTLs, where genetic variants were associated with acetylation variation, independently of disease. This was followed by downstream analyses for correlation to gene expression and overlap with sub-threshold signals from published LV-related GWAS datasets. RNA-seq was performed so as to generate matched tissue transcriptomes. Hi-C and HiChIP were performed for 2 independent control (NF) LV to generate a reference cardiac chromatin interactome. (B) Heatmap showing 3,897 differential H3K27-acetylation loci (FDR<0.05, fold-change ≥ 1.3; Wilcoxon Signed-Rank test), and top 10 transcription factor (TF) binding motifs enriched from the loci of hypo- and hyperacetylated peaks, respectively.