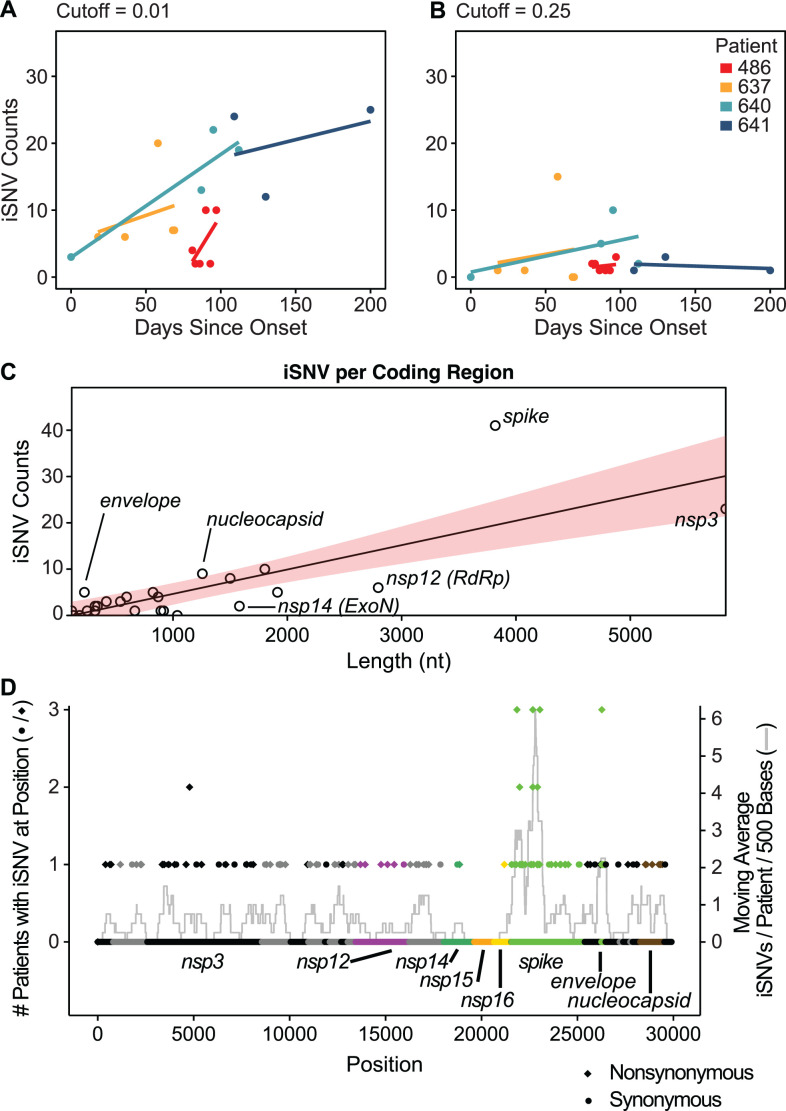
Fig 4.
iSNV accumulation over time and by the coding region. (A) iSNV accumulation across patients, shown over the full duration of sampled infection. Linear regression lines were fitted for a cutoff of 0.01 (iSNV found between 1% and 99% prevalence). (B) iSNV accumulation for a cutoff of 0.25 (25%–75% prevalence). (C) Number of iSNVs by the coding region showing length as a nucleotide on the x-axis and iSNV counts on the y-axis. Shaded region indicates the 95% CI for linear regression of coding region length to iSNV counts. (D) Occurrence of iSNVs throughout the genome by position. The left axis indicates the number of patients with any iSNV in a given position, indicated by either circles or diamonds. Circles indicate that the iSNV was synonymous, and diamonds indicate that the iSNV was nonsynonymous. The right axis indicates the moving average centered on each position spanning a window of 500 bases. Units for the moving average are average iSNVs per patient per 500 bases. Coding regions are colored in alternating black or gray, with the regions of interest labeled and colored.