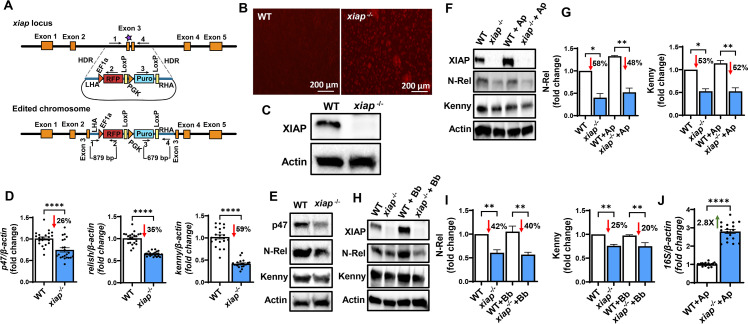
Fig 2.
CRISPR-Cas9 xiap editing in ISE6 cells. (A) Schematic representation of the xiap locus and the donor construct. The orange boxes represent the five exons of xiap with sgRNA binding and the Cas9 cleavage site on exon 3 (purple star). The donor construct contains the red fluorescent protein (RFP) and puromycin cassette (RFP-Puro) along with the DNA fragments of ~600 bp in length, homologous to the xiap locus, flanking the Cas9 cleavage site on the 5′ and 3′ ends for HDR. The arrows with numbers 1–4 represent the primers for the amplification analysis. (B and C) Editing was confirmed via fluorescence microscopy (B) and western blotting (C). (D and E) Functional disruption of xiap impaired the expression of molecules associated with the IMD pathway at both transcriptional (D) and translational levels (E). (F–I) 3 × 106 wild-type (WT) and xiap-/- cells were plated in a 6-well plate and stimulated with Anaplasma phagocytophilum [multiplicity of infection (MOI) 50] or Borrelia burgdorferi (MOI 50) for 15 minutes. Disruption of XIAP signaling impairs Kenny accumulation and Relish cleavage in response to (F and G) A. phagocytophilum infection or (H and I) B. burgdorferi stimulation. For data normalization, Kenny and N-Rel band densities were normalized to actin, and values were divided by the uninfected WT control densitometry. Western blot images are a representative image of at least two independent experiments. (J) WT and xiap-/- cells were infected with A. phagocytophilum (MOI 50). ISE6 cells were harvested after 72 hours of infection. The A. phagocytophilum 16S rRNA transcript was quantified by qRT-PCR, and the expression data were normalized to I. scapularis β-actin. The qRT-PCR data show the combination of three independent experiments. Results are represented as a mean ± SEM. Statistical significance was evaluated by unpaired t test with Welch’s correction (D and J) and one-way ANOVA with post hoc Tukey test (G and I). *P < 0.05; **P < 0.01; and ****P < 0.0001.