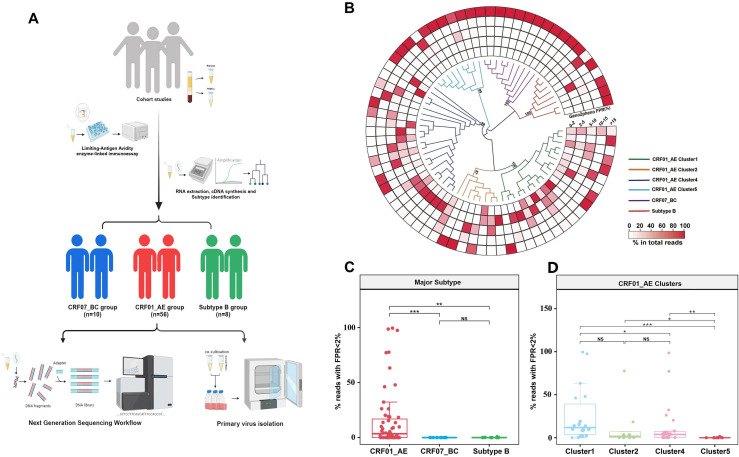
Fig 2.
Comparison of the predicted frequency of X-using strains among different HIV-1 sub-types and CRF01_AE clusters by next-generation sequencing. (A) Overview of the study participants. In the initial sample set; a total of 74 samples successfully met the screening criteria. This subset comprised 56 samples with pure CRF01_AE, 10 samples with CRF07_BC, and 8 samples with B sub-type infections. Next-generation sequencing was performed on plasma samples obtained from this selected population, and their peripheral blood mononuclear cells (PBMCs) underwent primary virus isolation. (B) Phylogenetic relationship of recently infected individuals in two cohort studies, including those infected with CRF01_AE clusters, CRF07_BC, and sub-type B viruses. The maximum likelihood tree was constructed using the most frequent haplotype sequences under the GTR + I + G site substitution model. Different colors represent sub-type sequences. (C and D) Heatmap depicting the frequency distribution of Geno2-pheno predicted false-positive rate (FPR) values in next-generation sequencing reads for each sample. The proportion of next-generation sequencing reads with Geno2pheno FPR of <2% in each sample was determined. Each dot indicates the frequency from a single individual. Two-tailed Mann-Whitney U test was employed to determine the statistical difference. ***P < 0.001, *P < 0.05, **P < 0.01, ***P < 0.001. NS, not statistically significant.